8IMN
 
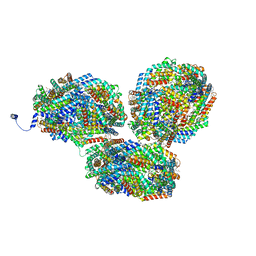 | | Rt1I-Rt1II, Rt2'I-Rt2'II, Rt3I-Rt3II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster F) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMM
 
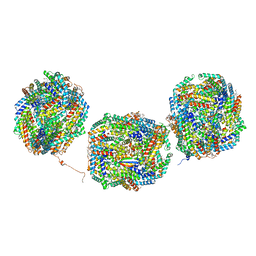 | | Rs2'I-Rs2'II, Rs1'I-Rs1'II, Rb'I-Rb'II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster E) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMO
 
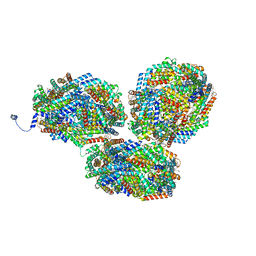 | | Rt1'I-Rt1'II, Rt2I-Rt2II, Rt3'I-Rt3'II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster G) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
7S3D
 
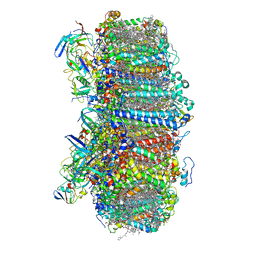 | | Structure of photosystem I with bound ferredoxin from Synechococcus sp. PCC 7335 acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2Fe-2S ferredoxin-type domain-containing protein, ... | | Authors: | Gisriel, C.J, Flesher, D.A, Shen, G, Wang, J, Ho, M, Brudvig, G.W, Bryant, D.A. | | Deposit date: | 2021-09-05 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structure of a photosystem I-ferredoxin complex from a marine cyanobacterium provides insights into far-red light photoacclimation.
J.Biol.Chem., 298, 2021
|
|
6PNJ
 
 | | Structure of Photosystem I Acclimated to Far-red Light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Gisriel, C.J, Shen, G, Kurashov, V, Ho, M, Zhang, S, Williams, D, Golbeck, J.H, Fromme, P, Bryant, D.A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | The structure of Photosystem I acclimated to far-red light illuminates an ecologically important acclimation process in photosynthesis
Sci Adv, 6, 2020
|
|
7TPR
 
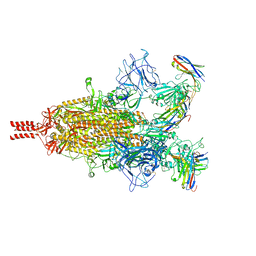 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
4X41
 
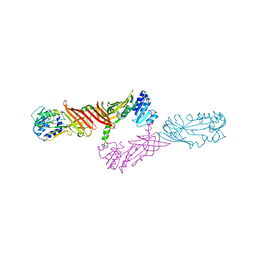 | | Crystal Structure of Protein Arginine Methyltransferase PRMT8 | | Descriptor: | Protein arginine N-methyltransferase 8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lee, W.C, Ho, M.C. | | Deposit date: | 2014-12-02 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Protein Arginine Methyltransferase 8: Tetrameric Structure and Protein Substrate Specificity
Biochemistry, 54, 2015
|
|
7BSJ
 
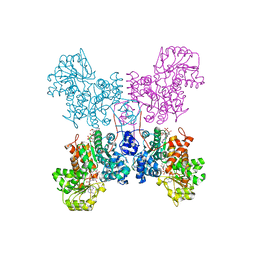 | | Crystal structure of human ME2 R484W | | Descriptor: | FUMARIC ACID, MAGNESIUM ION, NAD-dependent malic enzyme, ... | | Authors: | Chen, W.L, Tai, S.C, Hung, H.C, Ho, M.C. | | Deposit date: | 2020-03-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Single nucleotide variants lead to dysregulation of the human mitochondrial NAD(P) + -dependent malic enzyme.
Iscience, 24, 2021
|
|
3LMD
 
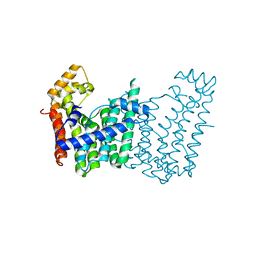 | | Crystal structure of geranylgeranyl pyrophosphate synthase from corynebacterium glutamicum atcc 13032 | | Descriptor: | Geranylgeranyl pyrophosphate synthase | | Authors: | Patskovsky, Y, Ho, M, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Corynebacterium Glutamicum
To be Published
|
|
7BSL
 
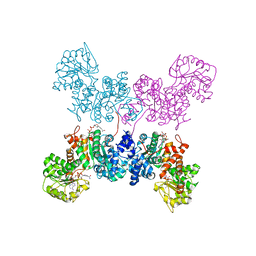 | | Crystal Structure of human ME2 R67A mutant | | Descriptor: | NAD-dependent malic enzyme, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, W.L, Tai, S.C, Hung, H.C, Ho, M.C. | | Deposit date: | 2020-03-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Single nucleotide variants lead to dysregulation of the human mitochondrial NAD(P) + -dependent malic enzyme.
Iscience, 24, 2021
|
|
7BSK
 
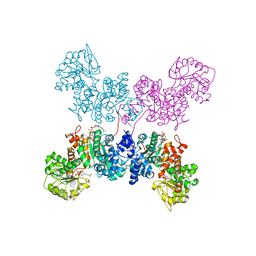 | | Crystal structure of human ME2 R67Q mutant | | Descriptor: | NAD-dependent malic enzyme, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, W.L, Tai, S.C, Hung, H.C, Ho, M.C. | | Deposit date: | 2020-03-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Single nucleotide variants lead to dysregulation of the human mitochondrial NAD(P) + -dependent malic enzyme.
Iscience, 24, 2021
|
|
3WRG
 
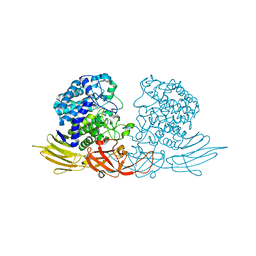 | | The complex structure of HypBA1 with L-arabinose | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRF
 
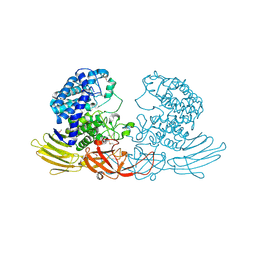 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
5YQ4
 
 | |
7YUH
 
 | | MtaLon-Apo for the spiral oligomers of trimer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUU
 
 | | MtaLon-ADP for the spiral oligomers of trimer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUV
 
 | | MtaLon-ADP for the spiral oligomers of tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUP
 
 | | MtaLon-Apo for the spiral oligomers of pentamer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUT
 
 | | MtaLon-Apo for the spiral oligomers of hexamer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUX
 
 | | MtaLon-ADP for the spiral oligomers of hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-18 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUW
 
 | | MtaLon-ADP for the spiral oligomers of pentamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-18 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUM
 
 | | MtaLon-Apo for the spiral oligomers of tetramer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
8GYK
 
 | | CryoEM structure of the RAD51_ADP filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair protein RAD51 homolog 1, MAGNESIUM ION | | Authors: | Miki, Y, Luo, S.C, Ho, M.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | A RAD51-ADP double filament structure unveils the mechanism of filament dynamics in homologous recombination.
Nat Commun, 14, 2023
|
|
8JC7
 
 | | Cryo-EM structure of Vibrio campbellii alpha-hemolysin | | Descriptor: | CALCIUM ION, Hemolysin, POTASSIUM ION | | Authors: | Wang, C.H, Yeh, M.K, Ho, M.C, Lin, S.M. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Structural basis for calcium-stimulating pore formation of Vibrio alpha-hemolysin.
Nat Commun, 14, 2023
|
|
4U3A
 
 | | Crystal structure of CtCel5E | | Descriptor: | Endoglucanase H | | Authors: | Yuan, S.F, Liang, P.H, Ho, M.C. | | Deposit date: | 2014-07-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Biochemical Characterization and Structural Analysis of a Bifunctional Cellulase/Xylanase from Clostridium thermocellum
J.Biol.Chem., 290, 2015
|
|
