2Z4H
 
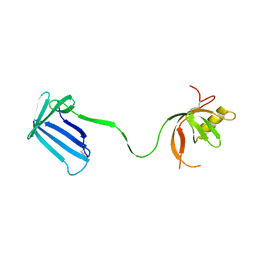 | | Crystal structure of the Cpx pathway activator NlpE from Escherichia coli | | Descriptor: | Copper homeostasis protein cutF, SULFATE ION | | Authors: | Hirano, Y, Hossain, M.M, Takeda, K, Tokuda, H, Miki, K. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-04 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of the Cpx Pathway Activator NlpE on the Outer Membrane of Escherichia coli
Structure, 15, 2007
|
|
3X34
 
 | | Crystal structure of the reduced form of the solubilized domain of porcine cytochrome b5 in form 1 crystal | | Descriptor: | CALCIUM ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirano, Y, Kimura, S, Tamada, T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.76 Å) | | Cite: | High-resolution crystal structures of the solubilized domain of porcine cytochrome b5.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3X32
 
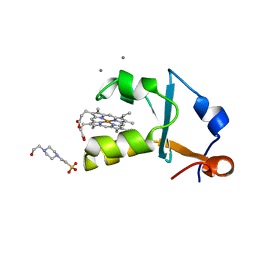 | | Crystal structure of the oxidized form of the solubilized domain of porcine cytochrome b5 in form 1 crystal | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Cytochrome b5, ... | | Authors: | Hirano, Y, Kimura, S, Tamada, T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | High-resolution crystal structures of the solubilized domain of porcine cytochrome b5.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3X35
 
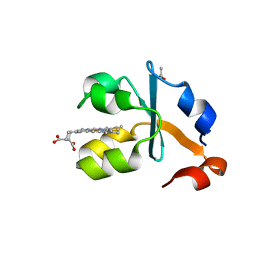 | | Crystal structure of the reduced form of the solubilized domain of porcine cytochrome b5 in form 2 crystal | | Descriptor: | ACETATE ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirano, Y, Kimura, S, Tamada, T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution crystal structures of the solubilized domain of porcine cytochrome b5.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3X33
 
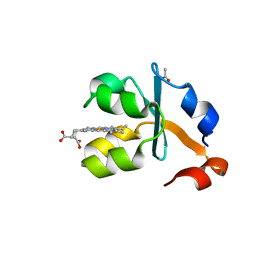 | | Crystal structure of the oxidized form of the solubilized domain of porcine cytochrome b5 in form 2 crystal | | Descriptor: | ACETATE ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirano, Y, Kimura, S, Tamada, T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | High-resolution crystal structures of the solubilized domain of porcine cytochrome b5.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6L46
 
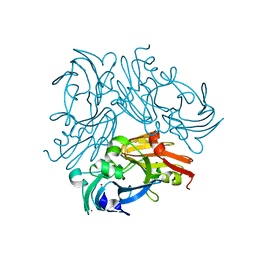 | | High-resolution neutron and X-ray joint refined structure of copper-containing nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Hirano, Y, Kusaka, K, Inoue, T, Tamada, T. | | Deposit date: | 2019-10-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.3 Å), X-RAY DIFFRACTION | | Cite: | High-resolution neutron crystallography visualizes an OH-bound resting state of a copper-containing nitrite reductase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6M3D
 
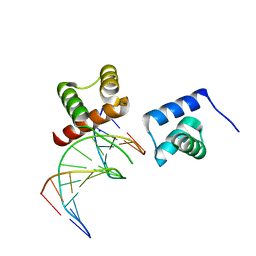 | | X-ray crystal structure of tandemly connected engrailed homeodomains (EHD) with R53A mutations and DNA complex | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*TP*AP*GP*GP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*TP*CP*CP*TP*AP*AP*TP*CP*C)-3'), SODIUM ION, ... | | Authors: | Sunami, T, Hirano, Y, Tamada, T, Kono, H. | | Deposit date: | 2020-03-03 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for designing an array of engrailed homeodomains.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5WQR
 
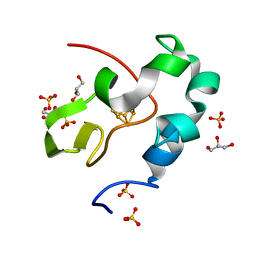 | | High resolution structure of high-potential iron-sulfur protein in the reduced state | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Ohno, H, Takeda, K, Niwa, S, Tsujinaka, T, Hanazono, Y, Hirano, Y, Miki, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic characterization of the high-potential iron-sulfur protein in the oxidized state at 0.8 angstrom resolution
PLoS ONE, 12, 2017
|
|
5WQQ
 
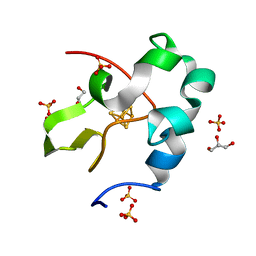 | | High resolution structure of high-potential iron-sulfur protein in the oxidized state | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Ohno, H, Takeda, K, Niwa, S, Tsujinaka, T, Hanazono, Y, Hirano, Y, Miki, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic characterization of the high-potential iron-sulfur protein in the oxidized state at 0.8 angstrom resolution
PLoS ONE, 12, 2017
|
|
4P9T
 
 | | Structure of the free form of the N-terminal VH1 domain of monomeric alpha-catenin | | Descriptor: | 1,2-ETHANEDIOL, Catenin alpha-2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shibahara, T, Hirano, Y, Hakoshima, T. | | Deposit date: | 2014-04-04 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the free form of the N-terminal VH1 domain of monomeric alpha-catenin.
Febs Lett., 589, 2015
|
|
7VOS
 
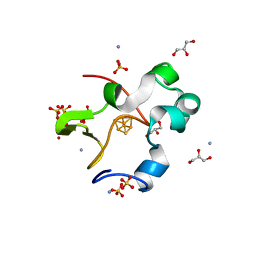 | | High-resolution neutron and X-ray joint refined structure of high-potential iron-sulfur protein in the oxidized state | | Descriptor: | AMMONIUM ION, GLYCEROL, High-potential iron-sulfur protein, ... | | Authors: | Hanazono, Y, Hirano, Y, Takeda, K, Kusaka, K, Tamada, T, Miki, K. | | Deposit date: | 2021-10-14 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (0.66 Å), X-RAY DIFFRACTION | | Cite: | Revisiting the concept of peptide bond planarity in an iron-sulfur protein by neutron structure analysis.
Sci Adv, 8, 2022
|
|
8IHY
 
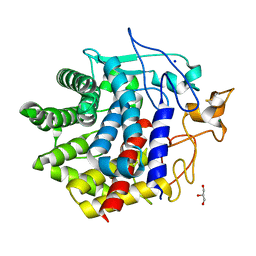 | | X-ray crystal structure of Q387E mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHW
 
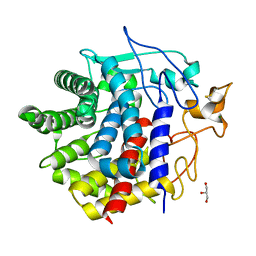 | | X-ray crystal structure of D43R mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHX
 
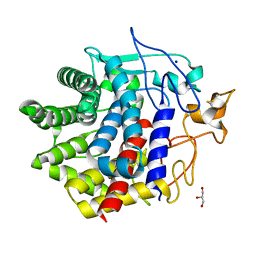 | | X-ray crystal structure of N372D mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
7CQY
 
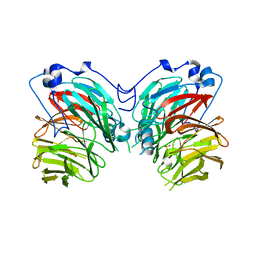 | |
6L8A
 
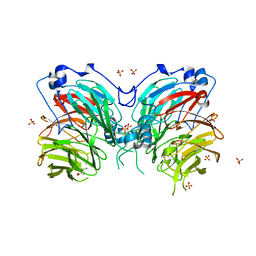 | | Tetrathionate hydrolase from Acidithiobacillus ferrooxidans | | Descriptor: | BETA-ALANINE, GLYCINE, SULFATE ION, ... | | Authors: | Tamada, T, Hirano, Y. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95004809 Å) | | Cite: | Reaction mechanism of tetrathionate hydrolysis based on the crystal structure of tetrathionate hydrolase from Acidithiobacillus ferrooxidans.
Protein Sci., 30, 2020
|
|
3W05
 
 | | Crystal structure of Oryza sativa DWARF14 (D14) in complex with PMSF | | Descriptor: | 1,2-ETHANEDIOL, Dwarf 88 esterase, phenylmethanesulfonic acid | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3W06
 
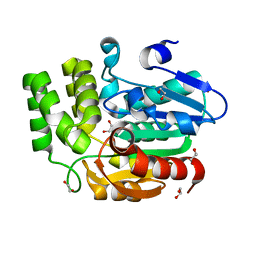 | | Crystal structure of Arabidopsis thaliana DWARF14 Like (AtD14L) | | Descriptor: | 1,2-ETHANEDIOL, Hydrolase, alpha/beta fold family protein | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3W04
 
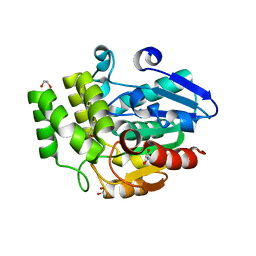 | | Crystal structure of Oryza sativa DWARF14 (D14) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3AAC
 
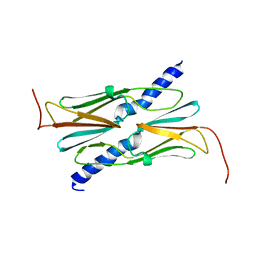 | | Small heat shock protein hsp14.0 with the mutations of I120F and I122F in the form II crystal | | Descriptor: | Putative uncharacterized protein ST1653 | | Authors: | Takeda, K, Hayashi, T, Abe, T, Hirano, Y, Hanazono, Y, Yohda, M, Miki, K. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dimer structure and conformational variability in the N-terminal region of an archaeal small heat shock protein, StHsp14.0
J.Struct.Biol., 174, 2011
|
|
3AAB
 
 | | Small heat shock protein hsp14.0 with the mutations of I120F and I122F in the form I crystal | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Putative uncharacterized protein ST1653 | | Authors: | Takeda, K, Hayashi, T, Abe, T, Hirano, Y, Hanazono, Y, Yohda, M, Miki, K. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Dimer structure and conformational variability in the N-terminal region of an archaeal small heat shock protein, StHsp14.0
J.Struct.Biol., 174, 2011
|
|
3WX2
 
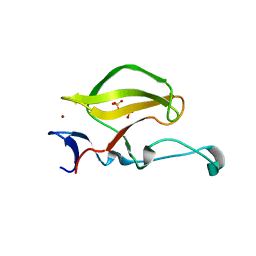 | | Mouse Cereblon thalidomide binding domain, native | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
3WX1
 
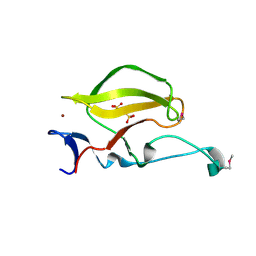 | | Mouse Cereblon thalidomide binding domain, selenomethionine derivative | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
3W6X
 
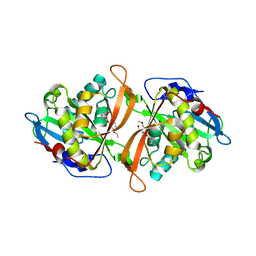 | | Yeast N-acetyltransferase Mpr1 in complex with CHOP | | Descriptor: | (4S)-4-hydroxy-L-proline, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W6S
 
 | | yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, MPR1 protein | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
