8A5B
 
 | | Crystal structure of human cathepsin L in complex with covalently bound MG-101 | | Descriptor: | Calpain Inhibitor I, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A4V
 
 | | Crystal structure of human cathepsin L with covalently bound E-64 | | Descriptor: | Cathepsin L, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
7AHA
 
 | | Structure of SARS-CoV-2 Main Protease bound to Maleate. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7QKC
 
 | | Crystal structure of human Cathepsin L after incubation with Sulfo-Calpeptin | | Descriptor: | Calpeptin, Cathepsin L, DI(HYDROXYETHYL)ETHER | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7AGA
 
 | | Structure of SARS-CoV-2 Main Protease bound to AT7519 | | Descriptor: | 3C-like proteinase, 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AR5
 
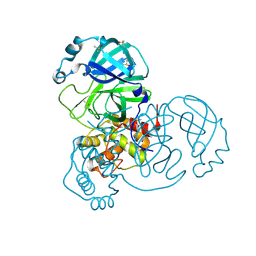 | | Structure of apo SARS-CoV-2 Main Protease with small beta angle, space group C2. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AMJ
 
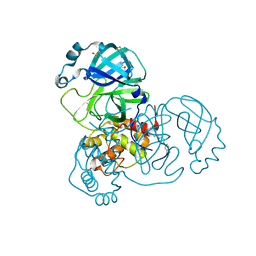 | | Structure of SARS-CoV-2 Main Protease bound to PD 168568. | | Descriptor: | (3~{S})-3-[2-[4-(3,4-dimethylphenyl)piperazin-1-yl]ethyl]-2,3-dihydroisoindol-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AR6
 
 | | Structure of apo SARS-CoV-2 Main Protease with large beta angle, space group C2. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6G8H
 
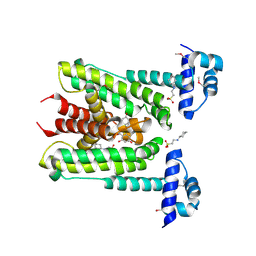 | | Flavonoid-responsive Regulator FrrA in complex with (R,S)-Naringenin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, NARINGENIN, R-naringenin, ... | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
6G87
 
 | | Flavonoid-responsive Regulator FrrA | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TetR/AcrR family transcriptional regulator | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-07 | | Release date: | 2019-04-24 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
6G8G
 
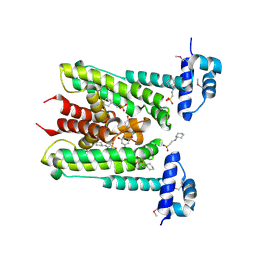 | | Flavonoid-responsive Regulator FrrA in complex with Genistein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GENISTEIN, TetR/AcrR family transcriptional regulator | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
2X3B
 
 | | AsaP1 inactive mutant E294A, an extracellular toxic zinc metalloendopeptidase | | Descriptor: | TOXIC EXTRACELLULAR ENDOPEPTIDASE, ZINC ION | | Authors: | Bogdanovic, X, Palm, G.J, Singh, R.K, Hinrichs, W. | | Deposit date: | 2010-01-22 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Evidence of Intramolecular Propeptide Inhibition of the Aspzincin Metalloendopeptidase Asap1.
FEBS Lett., 590, 2016
|
|
2X3C
 
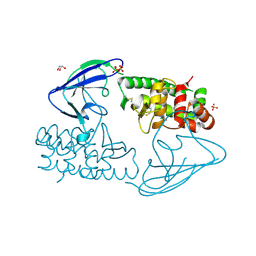 | | AsaP1 inactive mutant E294Q, an extracellular toxic zinc metalloendopeptidase | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Bogdanovic, X, Palm, G.J, Singh, R.K, Hinrichs, W. | | Deposit date: | 2010-01-22 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Evidence of Intramolecular Propeptide Inhibition of the Aspzincin Metalloendopeptidase Asap1.
FEBS Lett., 590, 2016
|
|
2X3A
 
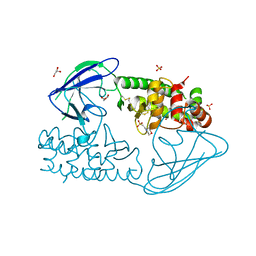 | | AsaP1 inactive mutant E294Q, an extracellular toxic zinc metalloendopeptidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Bogdanovic, X, Palm, G.J, Singh, R.K, Hinrichs, W. | | Deposit date: | 2010-01-22 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence of Intramolecular Propeptide Inhibition of the Aspzincin Metalloendopeptidase Asap1.
FEBS Lett., 590, 2016
|
|
2XT0
 
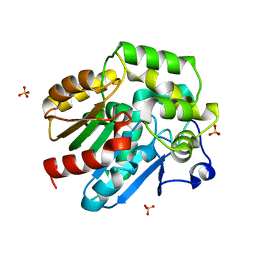 | | Dehalogenase DPpA from Plesiocystis pacifica SIR-I | | Descriptor: | HALOALKANE DEHALOGENASE, SULFATE ION | | Authors: | Bogdanovic, X, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-10-02 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cloning, Functional Expression, Biochemical Characterization, and Structural Analysis of a Haloalkane Dehalogenase from Plesiocystis Pacifica Sir-1.
Appl.Microbiol.Biotechnol., 91, 2011
|
|
7QKD
 
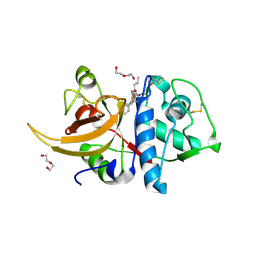 | | Crystal structure of human Cathepsin L in complex with covalently bound MG132 | | Descriptor: | ACETATE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
4AUX
 
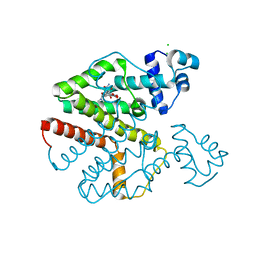 | | Tet repressor class D in complex with 9-nitrotetracycline | | Descriptor: | (4S,4aS,5aR,12aS)-4-(dimethylamino)-3,10,12,12a-tetrahydroxy-9-nitro-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Orth, P, Saenger, W, Hinrichs, W. | | Deposit date: | 2012-05-22 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tet Repressor Class D in Complex with 9-Nitrotetracycline and Magnesium
To be Published
|
|
6RN5
 
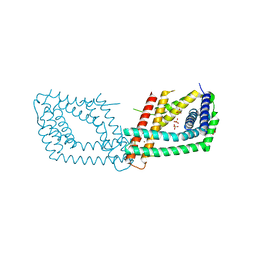 | | PptA from Streptomyces chartreusis | | Descriptor: | CARBONATE ION, CHAD domain protein, CHLORIDE ION, ... | | Authors: | Werten, S, Rustmeier, N.H, Hinrichs, W. | | Deposit date: | 2019-05-08 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | Structural and biochemical analysis of a phosin from Streptomyces chartreusis reveals a combined polyphosphate- and metal-binding fold.
Febs Lett., 593, 2019
|
|
2W3U
 
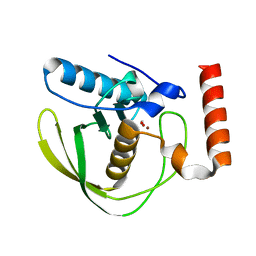 | | formate complex of the Ni-Form of E.coli deformylase | | Descriptor: | FORMIC ACID, NICKEL (II) ION, PEPTIDE DEFORMYLASE | | Authors: | Ngo, Y.H.T, Palm, G.J, Hinrichs, W. | | Deposit date: | 2008-11-14 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of the Ni(II) Complex of Escherichia Coli Peptide Deformylase and Suggestions on Deformylase Activities Depending on Different Metal(II) Centres.
J.Biol.Inorg.Chem., 15, 2010
|
|
2W3T
 
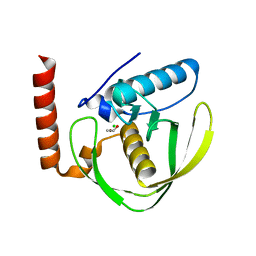 | | Chloro complex of the Ni-Form of E.coli deformylase | | Descriptor: | CHLORIDE ION, ETHANOL, NICKEL (II) ION, ... | | Authors: | Ngo, Y.H.T, Palm, G.J, Hinrichs, W. | | Deposit date: | 2008-11-14 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of the Ni(II) Complex of Escherichia Coli Peptide Deformylase and Suggestions on Deformylase Activities Depending on Different Metal(II) Centres.
J.Biol.Inorg.Chem., 15, 2010
|
|
2Y6Q
 
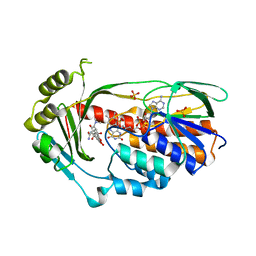 | | Structure of the TetX monooxygenase in complex with the substrate 7- Iodtetracycline | | Descriptor: | 7-IODOTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Wright, G.D, Hinrichs, W. | | Deposit date: | 2011-01-25 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
2Y6R
 
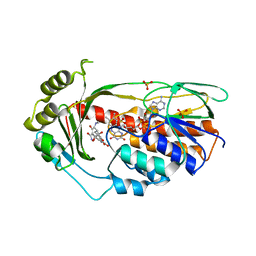 | | Structure of the TetX monooxygenase in complex with the substrate 7- chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Wright, G.D, Hinrichs, W. | | Deposit date: | 2011-01-25 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
7QCK
 
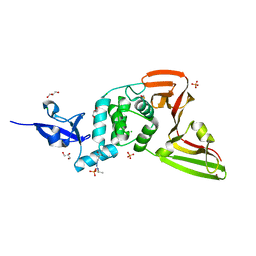 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCG
 
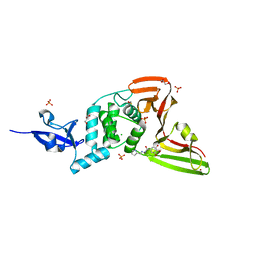 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
