4ZVR
 
 | |
4ZVP
 
 | |
4ZVO
 
 | |
4ZVQ
 
 | |
5JBM
 
 | |
1FRG
 
 | |
6CC0
 
 | |
6CBQ
 
 | |
3NM9
 
 | | HMGD(M13A)-DNA complex | | 分子名称: | DNA 5'-D(*G*GP*CP*GP*AP*TP*AP*TP*CP*GP*C)-3', High mobility group protein D | | 著者 | Churchill, M.E.A, Klass, J, Zoetewey, D.L. | | 登録日 | 2010-06-22 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural analysis of HMGD-DNA complexes reveals influence of intercalation on sequence selectivity and DNA bending.
J.Mol.Biol., 403, 2010
|
|
3SZT
 
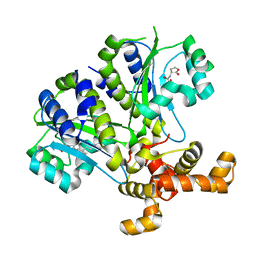 | | Quorum Sensing Control Repressor, QscR, Bound to N-3-oxo-dodecanoyl-L-Homoserine Lactone | | 分子名称: | N-3-OXO-DODECANOYL-L-HOMOSERINE LACTONE, Quorum-sensing control repressor, SODIUM ION | | 著者 | Churchill, M.E.A, Lintz, M.J. | | 登録日 | 2011-07-19 | | 公開日 | 2011-09-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4ZVT
 
 | |
4ZVS
 
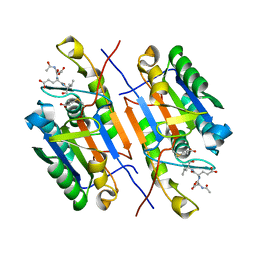 | |
4ZVU
 
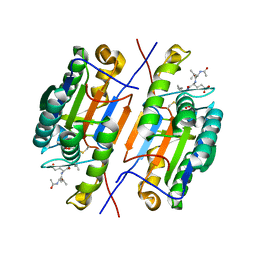 | |
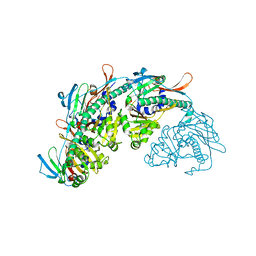
7SJ5
 
 | |
4EO5
 
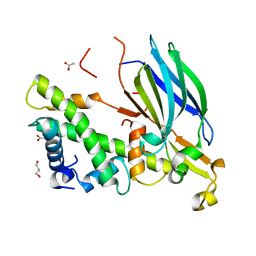 | | Yeast Asf1 bound to H3/H4G94P mutant | | 分子名称: | ACETATE ION, GLYCEROL, Histone H3.2, ... | | 著者 | Scorgie, J.K, Churchill, M.E. | | 登録日 | 2012-04-13 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The conformational flexibility of the C-terminus of histone H4 promotes histone octamer and nucleosome stability and yeast viability.
Epigenetics Chromatin, 5, 2012
|
|
1RO5
 
 | |
2PY0
 
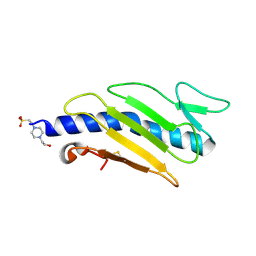 | | Crystal structure of Cs1 pilin chimera | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fimbrial protein | | 著者 | Kao, D.J, Churchill, M.E, Hodges, R.S. | | 登録日 | 2007-05-14 | | 公開日 | 2007-11-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Animal Protection and Structural Studies of a Consensus Sequence Vaccine Targeting the Receptor Binding Domain of the Type IV Pilus of Pseudomonas aeruginosa.
J.Mol.Biol., 374, 2007
|
|
1EG2
 
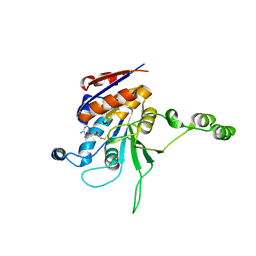 | | CRYSTAL STRUCTURE OF RHODOBACTER SPHEROIDES (N6 ADENOSINE) METHYLTRANSFERASE (M.RSRI) | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MODIFICATION METHYLASE RSRI | | 著者 | Scavetta, R.D, Thomas, C.B, Walsh, M.A, Szegedi, S, Joachimiak, A, Gumport, R.I, Churchill, M.E.A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2000-02-11 | | 公開日 | 2000-10-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases.
Nucleic Acids Res., 28, 2000
|
|
8DEI
 
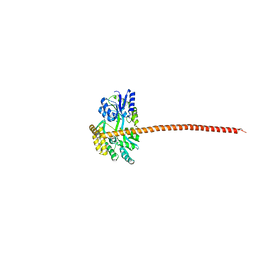 | | Structure of the Cac1 KER domain | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Maltodextrin-binding protein,Chromatin assembly factor 1 subunit p90 fusion, ... | | 著者 | Rosas, R, Churchill, M.E.A. | | 登録日 | 2022-06-20 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | A novel single alpha-helix DNA-binding domain in CAF-1 promotes gene silencing and DNA damage survival through tetrasome-length DNA selectivity and spacer function.
Elife, 12, 2023
|
|
4QR9
 
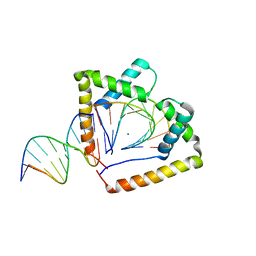 | | Crystal structure of two HMGB1 Box A domains cooperating to underwind and kink a DNA | | 分子名称: | DNA (5'-D(*AP*TP*AP*TP*CP*GP*AP*TP*AP*T)-3'), High mobility group protein B1, MAGNESIUM ION | | 著者 | Sanchez-Giraldo, R, Acosta-Reyes, F.J, Malarkey, C.S, Saperas, N, Churchill, M.E.A, Campos, J.L. | | 登録日 | 2014-06-30 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Two high-mobility group box domains act together to underwind and kink DNA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2C7A
 
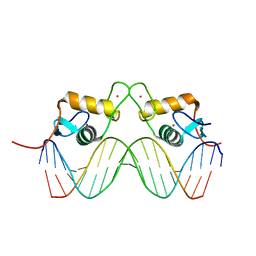 | | STRUCTURE OF THE PROGESTERONE RECEPTOR-DNA COMPLEX | | 分子名称: | 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*AP*AP *CP*TP*GP*TP*TP*CP*TP*G)-3', 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*TP *TP*TP*GP*TP*TP*CP*TP*G)-3', PROGESTERONE RECEPTOR, ... | | 著者 | Roemer, S.C, Donham, D.C, Sherman, L, Pon, V.H, Edwards, D.P, Churchill, M.E.A. | | 登録日 | 2005-11-19 | | 公開日 | 2006-08-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of the Progesterone Receptor-Deoxyribonucleic Acid Complex: Novel Interactions Required for Binding to Half-Site Response Elements.
Mol.Endocrinol., 20, 2006
|
|
3FGH
 
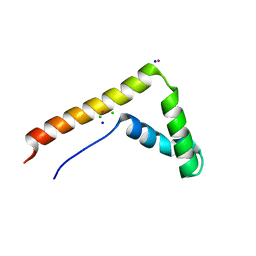 | | Human mitochondrial transcription factor A box B | | 分子名称: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | 著者 | Gangelhoff, T.A, Mungalachetty, P, Nix, J, Churchill, M.E.A. | | 登録日 | 2008-12-06 | | 公開日 | 2009-04-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A
Nucleic Acids Res., 37, 2009
|
|
2HUE
 
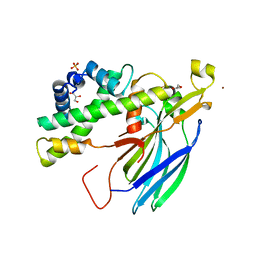 | | Structure of the H3-H4 chaperone Asf1 bound to histones H3 and H4 | | 分子名称: | Anti-silencing protein 1, GLYCEROL, Histone H3, ... | | 著者 | English, C.M, Churchill, M.E.A, Tyler, J.K. | | 登録日 | 2006-07-26 | | 公開日 | 2006-11-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for the histone chaperone activity of asf1.
Cell(Cambridge,Mass.), 127, 2006
|
|
1NW7
 
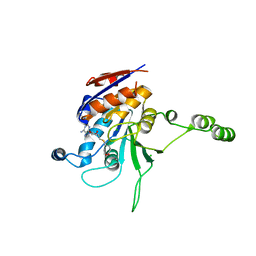 | | Structure of the beta class N6-adenine DNA methyltransferase RsrI bound to S-ADENOSYL-L-HOMOCYSTEINE | | 分子名称: | CHLORIDE ION, MODIFICATION METHYLASE RSRI, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | 登録日 | 2003-02-05 | | 公開日 | 2003-07-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
1NW6
 
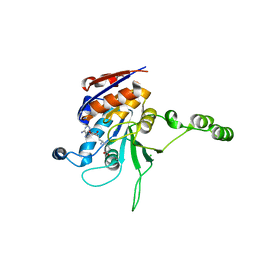 | | Structure of the beta class N6-adenine DNA methyltransferase RsrI bound to sinefungin | | 分子名称: | CHLORIDE ION, MODIFICATION METHYLASE RSRI, SINEFUNGIN | | 著者 | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | 登録日 | 2003-02-05 | | 公開日 | 2003-07-29 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
