4P16
 
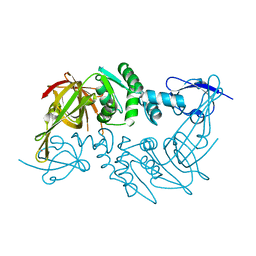 | | Crystal structure of the papain-like protease of Middle-East Respiratory Syndrome coronavirus | | Descriptor: | ORF1a, ZINC ION | | Authors: | Lei, J, Mesters, J.R, Ma, Q, Hilgenfeld, R. | | Deposit date: | 2014-02-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features.
Antiviral Res., 109C, 2014
|
|
6FV1
 
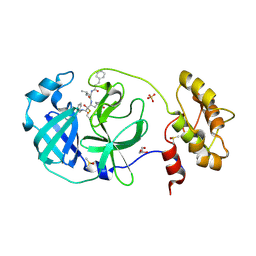 | | Structure of human coronavirus NL63 main protease in complex with the alpha-ketoamide (S)-N-((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)-2-cinnamamido-4-methylpentanamide (cinnamoyl-leucine-GlnLactam-CO-CO-NH-benzyl) | | Descriptor: | (2~{S})-4-methyl-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication Structure-based design, synthesis, and activity assessment.
J.Med.Chem., 2020
|
|
6FV2
 
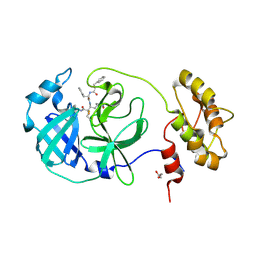 | |
1LK9
 
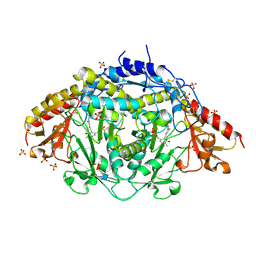 | | The Three-dimensional Structure of Alliinase from Garlic | | Descriptor: | 2-AMINO-ACRYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kuettner, E.B, Hilgenfeld, R, Weiss, M.S. | | Deposit date: | 2002-04-24 | | Release date: | 2002-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The active principle of garlic at atomic resolution
J.Biol.Chem., 277, 2002
|
|
1P9S
 
 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Replicase polyprotein 1ab | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
1EXM
 
 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS ELONGATION FACTOR TU (EF-TU) IN COMPLEX WITH THE GTP ANALOGUE GPPNHP. | | Descriptor: | ELONGATION FACTOR TU (EF-TU), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hogg, T, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2000-05-03 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the GTPase Mechanism of EF-Tu from Structural Studies
The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions, 28, 2000
|
|
1P9U
 
 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHQ-VNSTLQ-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION, ... | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
4K3A
 
 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | SULFATE ION, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K35
 
 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5HOL
 
 | |
5HIH
 
 | |
3O5Z
 
 | | Crystal structure of the SH3 domain from p85beta subunit of phosphoinositide 3-kinase (PI3K) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Chen, S, Xiao, Y, Ponnusamy, R, Tan, J, Lei, J, Hilgenfeld, R. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | X-ray structure of the SH3 domain of the phosphoinositide 3-kinase p85 beta subunit
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2XCU
 
 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6YXJ
 
 | |
3P31
 
 | |
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
2J98
 
 | | Human coronavirus 229E non structural protein 9 cys69ala mutant (Nsp9) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, REPLICASE POLYPROTEIN 1AB | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
2J97
 
 | | Human coronavirus 229E non structural protein 9 (Nsp9) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, REPLICASE POLYPROTEIN 1AB, SULFATE ION | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
8AIQ
 
 | | Mpro of SARS COV-2 in complex with the MG-87 inhibitor | | Descriptor: | CHLORIDE ION, Replicase polyprotein 1ab, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{S},3~{R})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Main Protease SARS-COV-2 in complex with the inhibitor MG-87
To Be Published
|
|
8AIV
 
 | | Mpro of SARS COV-2 in complex with the MG-100 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Main Protease SARS-COV-2 in complex with the inhibitor MG-100
To Be Published
|
|
8AIU
 
 | | Mpro of SARS COV-2 in complex with the MG-97 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Main Protease SARS-COV-2 in complex with the inhibitor MG-97
To Be Published
|
|
8AJ0
 
 | | Mpro of SARS COV-2 in complex with the RK-90 inhibitor | | Descriptor: | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(3-phenylpropanoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Main Protease SARS-COV-2 in complex with the inhibitor RK-90
To Be Published
|
|
8AIZ
 
 | | Mpro of SARS-CoV-2 in complex with the RK-68 inhibitor | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-3-cyclopropyl-2-[2-oxidanylidene-3-(2-phenylethanoylamino)pyridin-1-yl]propanoyl]amino]-~{N}-methyl-2-oxidanyl-4-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butanamide, CHLORIDE ION, Replicase polyprotein 1ab | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Main Protease SARS-CoV-2 in complex with the inhibitor RK-68
To Be Published
|
|
8A4T
 
 | | crystal structures of diastereomer (S,S,S)-13b (13b-K) in complex with the SARS-CoV-2 Mpro | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
8A4Q
 
 | | crystal structures of diastereomer (R,S,S)-13b (13b-H) in complex with the SARS-CoV-2 Mpro. | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
