2JCD
 
 | | Structure of the N-oxygenase AurF from Streptomyces thioluteus | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, N-OXIDASE | | Authors: | Zocher, G.E, Winkler, R, Hertweck, C, Schulz, G.E. | | Deposit date: | 2006-12-21 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure and Action of the N-Oxygenase Aurf from Streptomyces Thioluteus.
J.Mol.Biol., 373, 2007
|
|
5LVU
 
 | | XiaF (apo) from Streptomyces sp. | | Descriptor: | SULFATE ION, XiaF protein | | Authors: | Kugel, S, Baunach, M, Baer, P, Ishida-Ito, M, Sundaram, S, Xu, Z, Groll, M, Hertweck, C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cryptic indole hydroxylation by a non-canonical terpenoid cyclase parallels bacterial xenobiotic detoxification.
Nat Commun, 8, 2017
|
|
7BR0
 
 | | Crystal structure of AclR, a thioredoxin oxidoreductase fold protein carrying the CXXH catalytic motif | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyr_redox_2 domain-containing protein | | Authors: | Hara, K, Hashimoto, H, Maeda, N, Watanabe, K, Hertweck, C, Tsunematsu, Y. | | Deposit date: | 2020-03-26 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Specialized Flavoprotein Promotes Sulfur Migration and Spiroaminal Formation in Aspirochlorine Biosynthesis.
J.Am.Chem.Soc., 143, 2021
|
|
7ZHE
 
 | | Crystal structure of CtaZ from Ruminiclostridium cellulolyticum | | Descriptor: | GLYCEROL, SODIUM ION, Transcription activator effector binding | | Authors: | Gude, F, Molloy, E.M, Horch, T, Dell, M, Dunbar, K.L, Krabbe, J, Groll, M, Hertweck, C. | | Deposit date: | 2022-04-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Specialized Polythioamide-Binding Protein Confers Antibiotic Self-Resistance in Anaerobic Bacteria.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7ZHD
 
 | | Crystal structure of CtaZ in complex with Closthioamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, Transcription activator effector binding, ... | | Authors: | Gude, F, Molloy, E.M, Horch, T, Dell, M, Dunbar, K.L, Krabbe, J, Groll, M, Hertweck, C. | | Deposit date: | 2022-04-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Specialized Polythioamide-Binding Protein Confers Antibiotic Self-Resistance in Anaerobic Bacteria.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4NTC
 
 | | Crystal structure of GliT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GliT | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NTE
 
 | | Crystal structure of DepH | | Descriptor: | CHLORIDE ION, DepH, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NTD
 
 | | Crystal structure of HlmI | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3T6S
 
 | | Crystal structure of CerJ from Streptomyces tendae in Complex with CoA | | Descriptor: | COENZYME A, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-29 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3T8E
 
 | | Crystal structure of CerJ from Streptomyces tendae soaked with CerviK | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-08-01 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3T5Y
 
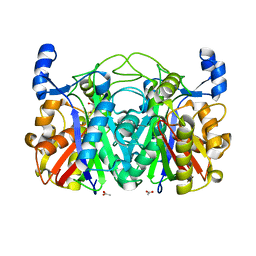 | | Crystal structure of CerJ from Streptomyces tendae - malonic acid covalently linked to the catalytic Cystein C116 | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
4KTU
 
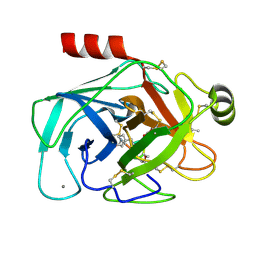 | | Bovine trypsin in complex with microviridin J at pH 6.5 | | Descriptor: | CALCIUM ION, Cationic trypsin, microviridin j | | Authors: | Quitterer, F, Groll, M, Hertweck, C, Dittmann, E. | | Deposit date: | 2013-05-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Harnessing the evolvability of tricyclic microviridins to dissect protease-inhibitor interactions.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4KTS
 
 | | Bovine trypsin in complex with microviridin J at pH 8.5 | | Descriptor: | CALCIUM ION, Cationic trypsin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Quitterer, F, Groll, M, Hertweck, C, Dittmann, E. | | Deposit date: | 2013-05-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Harnessing the evolvability of tricyclic microviridins to dissect protease-inhibitor interactions.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3P3Z
 
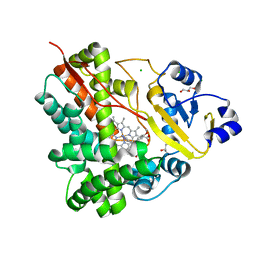 | | Crystal Structure of the Cytochrome P450 Monooxygenase AurH from Streptomyces Thioluteus in Complex with Ancymidol | | Descriptor: | (S)-cyclopropyl(4-methoxyphenyl)pyrimidin-5-ylmethanol, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | Deposit date: | 2010-10-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
3P3O
 
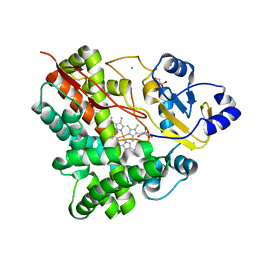 | | Crystal Structure of the Cytochrome P450 Monooxygenase AurH (ntermII) from Streptomyces Thioluteus | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | Deposit date: | 2010-10-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
3P3X
 
 | | Crystal Structure of the Cytochrome P450 Monooxygenase AurH (nterm-AurH-I) from Streptomyces Thioluteus | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | Deposit date: | 2010-10-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
3S3L
 
 | | Crystal Structure of CerJ from Streptomyces tendae | | Descriptor: | ACETATE ION, CHLORIDE ION, CerJ | | Authors: | Zocher, G, Hertweck, C, Bretschneider, T, Stehle, T. | | Deposit date: | 2011-05-18 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3P3L
 
 | | Crystal Structure of the Cytochrome P450 monooxygenase AurH (wildtype) from Streptomyces Thioluteus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | Deposit date: | 2010-10-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
7PCE
 
 | | BurG (apo): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ketol-acid reductoisomerase, PHOSPHATE ION | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCC
 
 | | BurG in complex with Mg2+ and NAD+ (holo): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCG
 
 | | BurG (holo) in complex with cyclopropane-1,1-dicarboxylate (7): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCM
 
 | | BurG (holo) in complex with (Z)-2,3-dihydroxy-6-methyl-hept-2-enoate (13): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-6-methyl-2,3-bis(oxidanyl)hept-2-enoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCO
 
 | | BurG E232Q mutant (holo) in complex with 2R,3R-2,3-dihydroxy-6-methyl-heptanoate (12): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R,3R)-6-methyl-2,3-bis(oxidanyl)heptanoic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCI
 
 | | BurG (holo) in complex with hydroxypyruvate-enol (8): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-2,3-bis(oxidanyl)prop-2-enoic acid, IMIDAZOLE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCT
 
 | | BurG E232Q mutant (holo) in complex with enol-oxalacetate (15): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
