3LCK
 
 | |
3LIA
 
 | |
3M72
 
 | |
3M74
 
 | |
3M7E
 
 | |
3M71
 
 | | Crystal Structure of Plant SLAC1 homolog TehA | | Descriptor: | Tellurite resistance protein tehA homolog, octyl beta-D-glucopyranoside | | Authors: | Chen, Y.-H, Hu, L, Punta, M, Bruni, R, Hillerich, B, Kloss, B, Rost, B, Love, J, Siegelbaum, S.A, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-03-16 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Homologue structure of the SLAC1 anion channel for closing stomata in leaves.
Nature, 467, 2010
|
|
3M7C
 
 | |
3M76
 
 | |
3M77
 
 | |
3M70
 
 | |
3M75
 
 | |
3M73
 
 | |
3M7B
 
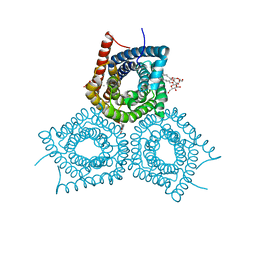 | |
3M7L
 
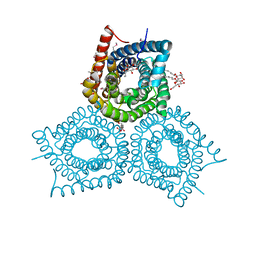 | |
3O1H
 
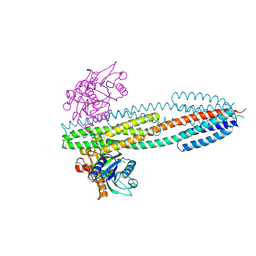 | |
3M78
 
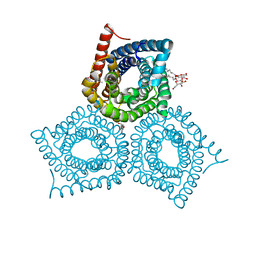 | |
3O1J
 
 | |
3O1I
 
 | |
1SCF
 
 | | HUMAN RECOMBINANT STEM CELL FACTOR | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, STEM CELL FACTOR | | Authors: | Jiang, X, Gurel, O, Langley, K.E, Hendrickson, W.A. | | Deposit date: | 1998-06-04 | | Release date: | 2000-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the active core of human stem cell factor and analysis of binding to its receptor kit.
EMBO J., 19, 2000
|
|
1T20
 
 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9-6I | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
1T22
 
 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9, orthorhombic crystal | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
1TEN
 
 | |
1TTU
 
 | | Crystal Structure of CSL bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3', 5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3', ... | | Authors: | Kovall, R.A, Hendrickson, W.A. | | Deposit date: | 2004-06-23 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA
Embo J., 23, 2004
|
|
1RZ8
 
 | | CRYSTAL STRUCTURE OF HUMAN ANTI-HIV-1 GP120-REACTIVE ANTIBODY 17B | | Descriptor: | Fab 17b heavy chain, Fab 17b light chain | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T1Z
 
 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9-6A | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
