3PRD
 
 | |
4RYN
 
 | | Crystal structure of BcTSPO, type1 monomer | | Descriptor: | CACODYLATE ION, DODECYL-ALPHA-D-MALTOSIDE, Integral membrane protein, ... | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
1IRK
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HUMAN INSULIN RECEPTOR | | Descriptor: | ETHYL MERCURY ION, INSULIN RECEPTOR TYROSINE KINASE DOMAIN | | Authors: | Hubbard, S.R, Wei, L, Ellis, L, Hendrickson, W.A. | | Deposit date: | 1995-01-02 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of the human insulin receptor.
Nature, 372, 1994
|
|
1CDH
 
 | |
1CDI
 
 | |
1NCJ
 
 | | N-CADHERIN, TWO-DOMAIN FRAGMENT | | Descriptor: | CALCIUM ION, PROTEIN (N-CADHERIN), URANYL (VI) ION | | Authors: | Tamura, K, Shan, W.-S, Hendrickson, W.A, Colman, D.R, Shapiro, L. | | Deposit date: | 1999-02-02 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-function analysis of cell adhesion by neural (N-) cadherin.
Neuron, 20, 1998
|
|
8D1M
 
 | |
4TKQ
 
 | | Native-SAD phasing for YetJ from Bacillus Subtilis | | Descriptor: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein YetJ | | Authors: | Liu, Q, Chang, Y, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8025 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4RML
 
 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in C2221 crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Latrophilin-3, MAGNESIUM ION | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
4RMK
 
 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in P65 crystal form | | Descriptor: | CALCIUM ION, Latrophilin-3 | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
4RYI
 
 | |
4WB8
 
 | | Crystal structure of human cAMP-dependent protein kinase A (catalytic alpha subunit), exon 1 deletion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1CD8
 
 | |
3LIB
 
 | |
1GC1
 
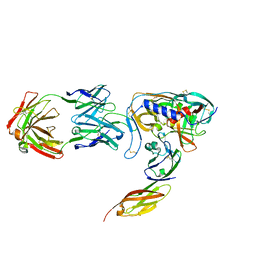 | | HIV-1 GP120 CORE COMPLEXED WITH CD4 AND A NEUTRALIZING HUMAN ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, CD4, ... | | Authors: | Kwong, P.D, Wyatt, R, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 1998-06-15 | | Release date: | 1998-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody.
Nature, 393, 1998
|
|
1HR3
 
 | |
1BDO
 
 | |
1AXM
 
 | | HEPARIN-LINKED BIOLOGICALLY-ACTIVE DIMER OF FIBROBLAST GROWTH FACTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | DiGabriele, A.D, Lax, I, Chen, D.I, Svahn, C.M, Jaye, M, Schlessinger, J, Hendrickson, W.A. | | Deposit date: | 1997-10-16 | | Release date: | 1998-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a heparin-linked biologically active dimer of fibroblast growth factor.
Nature, 393, 1998
|
|
1BUN
 
 | | STRUCTURE OF BETA2-BUNGAROTOXIN: POTASSIUM CHANNEL BINDING BY KUNITZ MODULES AND TARGETED PHOSPHOLIPASE ACTION | | Descriptor: | BETA2-BUNGAROTOXIN, SODIUM ION | | Authors: | Kwong, P.D, Mcdonald, N.Q, Sigler, P.B, Hendrickson, W.A. | | Deposit date: | 1995-10-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action.
Structure, 3, 1995
|
|
1RNH
 
 | | STRUCTURE OF RIBONUCLEASE H PHASED AT 2 ANGSTROMS RESOLUTION BY MAD ANALYSIS OF THE SELENOMETHIONYL PROTEIN | | Descriptor: | RIBONUCLEASE HI, SULFATE ION | | Authors: | Yang, W, Hendrickson, W.A, Crouch, R.J, Satow, Y. | | Deposit date: | 1990-07-11 | | Release date: | 1991-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein.
Science, 249, 1990
|
|
1NEU
 
 | | STRUCTURE OF MYELIN MEMBRANE ADHESION MOLECULE P0 | | Descriptor: | MYELIN P0 PROTEIN | | Authors: | Shapiro, L, Doyle, J.P, Hensley, P, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1996-09-24 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin.
Neuron, 17, 1996
|
|
1NCH
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCG
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCI
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, URANYL (VI) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
5TAQ
 
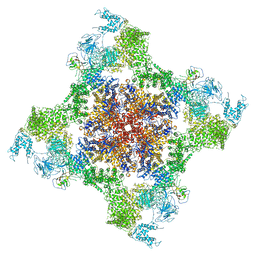 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 3&4) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
