6OI4
 
 | | RPN13 (19-132)-RPN2 (940-952) pY950-Ub complex | | Descriptor: | 1,2-ETHANEDIOL, 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, ... | | Authors: | Hemmis, C.W, Hill, C.P. | | Deposit date: | 2019-04-08 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Phosphorylation of Tyr-950 in the proteasome scaffolding protein RPN2 modulates its interaction with the ubiquitin receptor RPN13.
J.Biol.Chem., 294, 2019
|
|
5V1Y
 
 | | Crystal structure of the ternary RPN13 PRU-RPN2 (940-953)-ubiquitin complex | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Hemmis, C.W, VanderLinden, R.T, Yao, T, Robinson, H, Hill, C.P. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism.
J. Biol. Chem., 292, 2017
|
|
5V1Z
 
 | | Crystal structure of the RPN13 PRU-RPN2 (932-953)-ubiquitin complex | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Hemmis, C.W, VanderLinden, R.T, Yao, T, Robinson, H, Hill, C.P. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism.
J. Biol. Chem., 292, 2017
|
|
4WLR
 
 | | Crystal Structure of mUCH37-hRPN13 CTD-hUb complex | | Descriptor: | Polyubiquitin-B, Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
4WLP
 
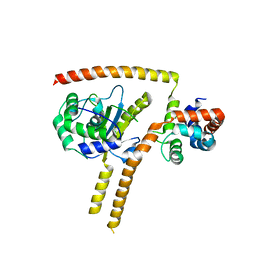 | | Crystal structure of UCH37-NFRKB Inhibited Deubiquitylating Complex | | Descriptor: | Nuclear factor related to kappa-B-binding protein, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
4WLQ
 
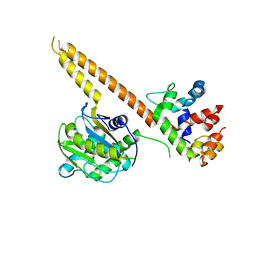 | | Crystal structure of mUCH37-hRPN13 CTD complex | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
