8CJJ
 
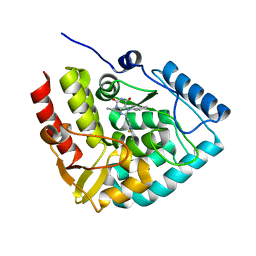 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-057 | | Descriptor: | 3-ethyl-7-(phenylmethyl)-8-(5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-2-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66415656 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CJN
 
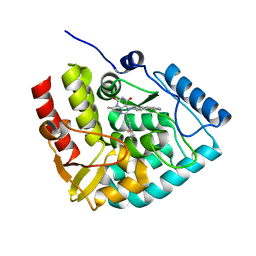 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-070 | | Descriptor: | 3-ethyl-7-[(4-phenylphenyl)methyl]-8-(5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-2-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68080938 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
3JZ7
 
 | |
2JJZ
 
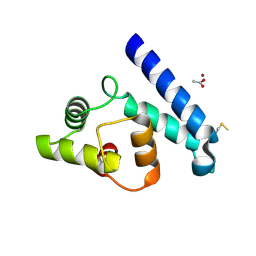 | | Crystal Structure of Human Iba2, orthorhombic crystal form | | Descriptor: | ACETATE ION, CHLORIDE ION, IONIZED CALCIUM-BINDING ADAPTER MOLECULE 2, ... | | Authors: | Schulze, J.O, Quedenau, C, Roske, Y, Turnbull, A, Mueller, U, Heinemann, U, Buessow, K. | | Deposit date: | 2008-05-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of Human Iba Proteins.
FEBS J., 275, 2008
|
|
1XCR
 
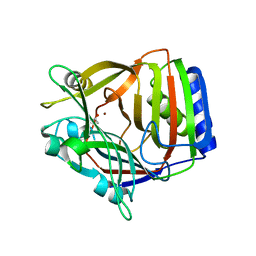 | | Crystal Structure of Longer Splice Variant of PTD012 from Homo sapiens reveals a novel Zinc-containing fold | | Descriptor: | ACETIC ACID, ZINC ION, hypothetical protein PTD012 | | Authors: | Manjasetty, B.A, Fieber-Erdmann, M, Roske, Y, Goetz, F, Buessow, K, Heinemann, U. | | Deposit date: | 2004-09-03 | | Release date: | 2005-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Homo sapiens PTD012 reveals a zinc-containing hydrolase fold
Protein Sci., 15, 2006
|
|
3KXC
 
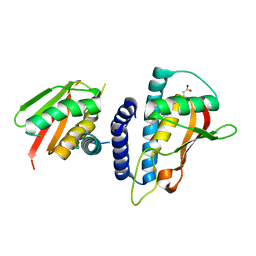 | | Mutant transport protein | | Descriptor: | PALMITIC ACID, Trafficking protein particle complex subunit 3, Trafficking protein particle complex subunit 6B | | Authors: | Kummel, D, Heinemann, U. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the self-palmitoylation activity of the transport protein particle component Bet3
Cell.Mol.Life Sci., 67, 2010
|
|
2OKN
 
 | | Crystal Strcture of Human Prolidase | | Descriptor: | HYDROGENPHOSPHATE ION, MANGANESE (II) ION, Xaa-Pro dipeptidase | | Authors: | Mueller, U, Niesen, F.H, Roske, Y, Goetz, F, Behlke, J, Buessow, K, Heinemann, U, Protein Structure Factory (PSF) | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Prolidase: The Molecular Basis of PD Disease.
To be Published
|
|
3O5N
 
 | | Tetrahydroquinoline carboxylates are potent inhibitors of the Shank PDZ domain, a putative target in autism disorders | | Descriptor: | (3aS,4R,9bR)-9-nitro-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-4,6-dicarboxylic acid, SH3 and multiple ankyrin repeat domains protein 3 | | Authors: | Saupe, J, Roske, Y, Schillinger, C, Kamdem, N, Radetzki, S, Diehl, A, Oschkinat, H, Krause, G, Heinemann, U, Rademann, J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery, structure-activity relationship studies, and crystal structure of nonpeptide inhibitors bound to the shank3 PDZ domain.
Chemmedchem, 6, 2011
|
|
1NEG
 
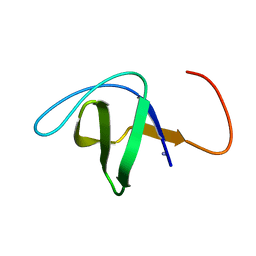 | | Crystal Structure Analysis of N-and C-terminal labeled SH3-domain of alpha-Chicken Spectrin | | Descriptor: | AZIDE ION, Spectrin alpha chain, brain | | Authors: | Mueller, U, Buessow, K, Diehl, A, Niesen, F.H, Nyarsik, L, Heinemann, U. | | Deposit date: | 2002-12-11 | | Release date: | 2003-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid purification and crystal structure analysis of a small protein carrying two terminal affinity tags
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
1ONI
 
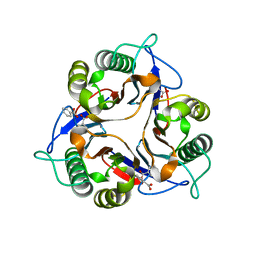 | | Crystal structure of a human p14.5, a translational inhibitor reveals different mode of ligand binding near the invariant residues of the Yjgf/UK114 protein family | | Descriptor: | 14.5 kDa translational inhibitor protein, BENZOIC ACID | | Authors: | Manjasetty, B.A, Delbrueck, H, Mueller, U, Erdmann, M.F, Heinemann, U. | | Deposit date: | 2003-02-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Homo sapiens protein hp14.5.
Proteins, 54, 2004
|
|
2RNT
 
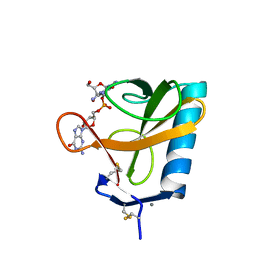 | | THREE-DIMENSIONAL STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH GUANYLYL-2(PRIME),5(PRIME)-GUANOSINE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, GUANYLYL-2',5'-PHOSPHOGUANOSINE, RIBONUCLEASE T1 | | Authors: | Saenger, W, Koepke, J, Maslowska, M, Heinemann, U. | | Deposit date: | 1988-07-06 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of ribonuclease T1 complexed with guanylyl-2',5'-guanosine at 1.8 A resolution.
J.Mol.Biol., 206, 1989
|
|
5IFW
 
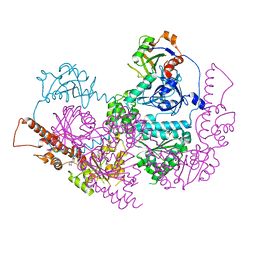 | |
5IFS
 
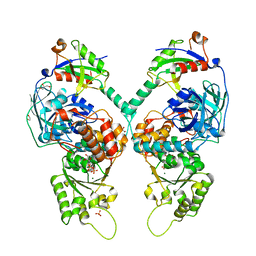 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | Deposit date: | 2016-02-26 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
2ONQ
 
 | |
2ON8
 
 | |
7NDH
 
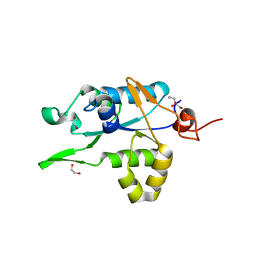 | | Crystal structure of ZC3H12C PIN domain | | Descriptor: | 1,2-ETHANEDIOL, Probable ribonuclease ZC3H12C, SODIUM ION | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
7NDJ
 
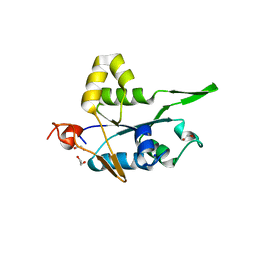 | |
7NDI
 
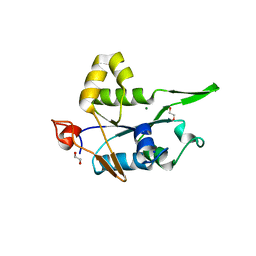 | | Crystal structure of ZC3H12C PIN domain with Mg2+ Ion | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable ribonuclease ZC3H12C, ... | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
7NDK
 
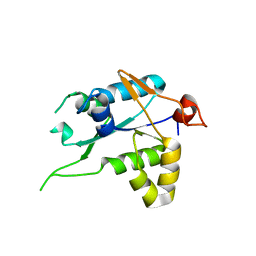 | | Crystal structure of ZC3H12C PIN catalytic mutant | | Descriptor: | Probable ribonuclease ZC3H12C, SODIUM ION | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
1MJC
 
 | |
4ZP3
 
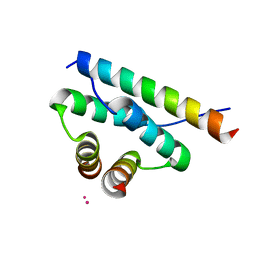 | | AKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA | | Descriptor: | A-kinase anchor protein 7 isoforms alpha and beta, CADMIUM ION, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Goetz, F, Roske, Y, Faelber, K, Zuehlke, K, Autenrieth, K, Kreuchwig, A, Krause, G, Herberg, F.W, Daumke, O, Heinemann, U, Klussmann, E. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | AKAP18:PKA-RII alpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA.
Biochem.J., 473, 2016
|
|
3PR6
 
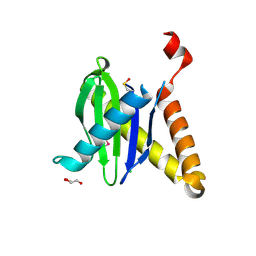 | |
5MR7
 
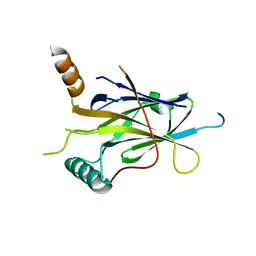 | |
5MPH
 
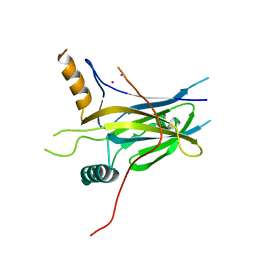 | | Structural Basis of Gene Regulation by the Grainyhead Transcription Factor Superfamily | | Descriptor: | Grainyhead-like protein 1 homolog, IODIDE ION | | Authors: | Ming, Q, Roske, Y, Schuetz, A, Walentin, K, Ibraimi, I, Schmidt-Ott, K.M, Heinemann, U. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.337 Å) | | Cite: | Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family.
Nucleic Acids Res., 46, 2018
|
|
5MPF
 
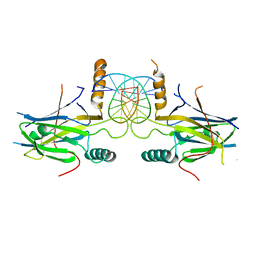 | | Structural Basis of Gene Regulation by the Grainyhead Transcription Factor Superfamily | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*CP*CP*GP*GP*TP*TP*TP*T)-3'), Grainyhead-like protein 1 homolog | | Authors: | Ming, Q, Roske, Y, Schuetz, A, Walentin, K, Ibraimi, I, Schmidt-Ott, K.M, Heinemann, U. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family.
Nucleic Acids Res., 46, 2018
|
|
