2D25
 
 | | C-C-A-G-G-C-M5C-T-G-G; HELICAL FINE STRUCTURE, HYDRATION, AND COMPARISON WITH C-C-A-G-G-C-C-T-G-G | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*(5CM)P*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Heinemann, U, Hahn, M. | | Deposit date: | 1991-04-23 | | Release date: | 1991-04-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | C-C-A-G-G-C-m5C-T-G-G. Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G.
J.Biol.Chem., 267, 1992
|
|
9DNA
 
 | |
1D26
 
 | |
1CGC
 
 | |
3ANA
 
 | |
1BD1
 
 | | CRYSTALLOGRAPHIC STUDY OF ONE TURN OF G/C-RICH B-DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G)-3'), TRIETHYLAMMONIUM ION | | Authors: | Heinemann, U. | | Deposit date: | 1989-08-16 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic study of one turn of G/C-rich B-DNA.
J.Mol.Biol., 210, 1989
|
|
6HD0
 
 | |
6HD3
 
 | | Common mode of remodeling AAA ATPases p97/CDC48 by their disassembly cofactors ASPL/PUX1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48 homolog A, PHOSPHATE ION | | Authors: | Heinemann, U, Roske, Y, Banchenko, S, Arumughan, A, Petrovic, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Common Mode of Remodeling AAA ATPases p97/CDC48 by Their Disassembling Cofactors ASPL/PUX1.
Structure, 27, 2019
|
|
1MAC
 
 | | CRYSTAL STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF BACILLUS MACERANS ENDO-1,3-1,4-BETA-GLUCANASE | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase.
J.Biol.Chem., 270, 1995
|
|
1MJC
 
 | |
5G27
 
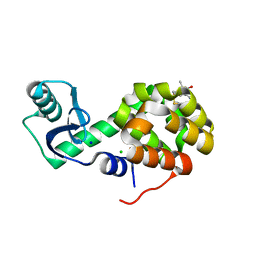 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at Room Temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ENDOLYSIN, ... | | Authors: | Gohlke, U, Consentius, P, Loll, B, Mueller, R, Kaupp, M, Heinemann, U, Risse, T. | | Deposit date: | 2016-04-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-Ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
4YEJ
 
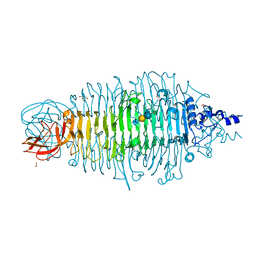 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 in complex with pentasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4YEL
 
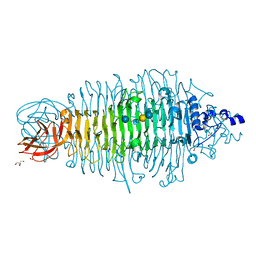 | | Tailspike protein double mutant D339A/E372A of E. coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
3ULJ
 
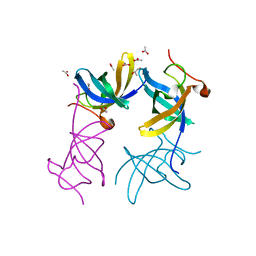 | | Crystal structure of apo Lin28B cold shock domain | | Descriptor: | ACETATE ION, GLYCEROL, Lin28b, ... | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2011-11-10 | | Release date: | 2012-08-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The Lin28 cold-shock domain remodels pre-let-7 microRNA.
Nucleic Acids Res., 40, 2012
|
|
9RNT
 
 | |
3KXC
 
 | | Mutant transport protein | | Descriptor: | PALMITIC ACID, Trafficking protein particle complex subunit 3, Trafficking protein particle complex subunit 6B | | Authors: | Kummel, D, Heinemann, U. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the self-palmitoylation activity of the transport protein particle component Bet3
Cell.Mol.Life Sci., 67, 2010
|
|
3RNT
 
 | | CRYSTAL STRUCTURE OF GUANOSINE-FREE RIBONUCLEASE T1, COMPLEXED WITH VANADATE(V), SUGGESTS CONFORMATIONAL CHANGE UPON SUBSTRATE BINDING | | Descriptor: | CALCIUM ION, RIBONUCLEASE T1, VANADATE ION | | Authors: | Kostrewa, D, Choe, H.-W, Heinemann, U, Saenger, W. | | Deposit date: | 1989-05-31 | | Release date: | 1989-10-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of guanosine-free ribonuclease T1, complexed with vanadate (V), suggests conformational change upon substrate binding.
Biochemistry, 28, 1989
|
|
2XKW
 
 | | LIGAND BINDING DOMAIN OF HUMAN PPAR-GAMMA IN COMPLEX WITH THE AGONIST PIOGLITAZONE | | Descriptor: | (5R)-5-{4-[2-(5-ethylpyridin-2-yl)ethoxy]benzyl}-1,3-thiazolidine-2,4-dione, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Mueller, J.J, Schupp, M, Unger, T, Kintscher, U, Heinemann, U. | | Deposit date: | 2010-07-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Binding Diversity of Pioglitazone by Peroxisome Proliferator-Activated Receptor-Gamma
To be Published
|
|
4AVZ
 
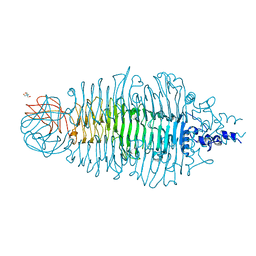 | | Tailspike protein mutant E372Q of E. coli bacteriophage HK620 | | Descriptor: | TAIL SPIKE PROTEIN, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Gohlke, U, Broeker, N.K, Mueller, J.J, Uetrecht, C, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
2XC1
 
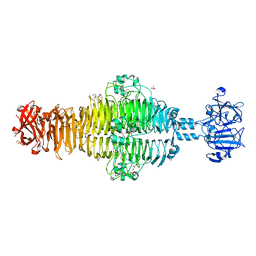 | | Full-length Tailspike Protein Mutant Y108W of Bacteriophage P22 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, ... | | Authors: | Mueller, J.J, Seul, A, Seckler, R, Heinemann, U. | | Deposit date: | 2010-04-15 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bacteriophage P22 Tailspike: Structure of the Complete Protein and Function of the Interdomain Linker
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ULW
 
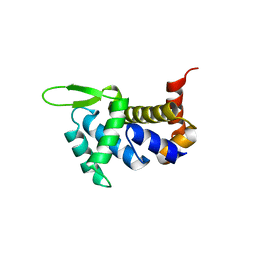 | |
3JZ7
 
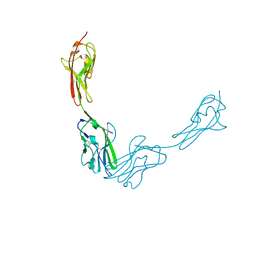 | |
1RNT
 
 | | RESTRAINED LEAST-SQUARES REFINEMENT OF THE CRYSTAL STRUCTURE OF THE RIBONUCLEASE T1(ASTERISK)2(PRIME)-GUANYLIC ACID COMPLEX AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 ISOZYME | | Authors: | Saenger, W, Arni, R, Heinemann, U, Tokuoka, R. | | Deposit date: | 1987-07-10 | | Release date: | 1987-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restrained Least-Squares Refinement of the Crystal Structure of the Ribonuclease T1(Asterisk)2(Prime)-Guanylic Acid Complex at 1.9 Angstroms Resolution
Acta Crystallogr.,Sect.B, 43, 1987
|
|
1LRA
 
 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
6F5Q
 
 | | Crystal Structure of KDM4D with GF026 ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(methylsulfonylamino)ethylamino]pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal Structure of KDM4D with GF026 ligand
To be published
|
|
