2WCM
 
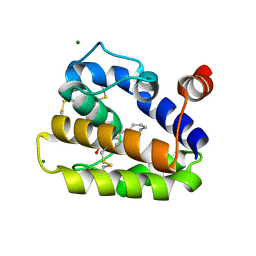 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (10E)-hexadecen-12-yn-1-ol | | Descriptor: | (10E)-hexadec-10-en-12-yn-1-ol, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WCK
 
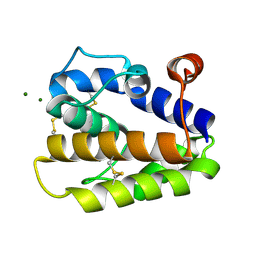 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) without ligand | | Descriptor: | GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WCL
 
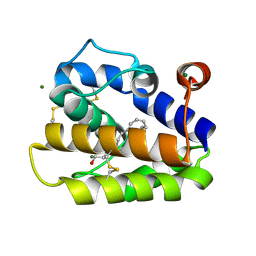 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (8E, 10Z)-hexadecadien-1-ol | | Descriptor: | (8E,10Z)-HEXADECA-8,10-DIEN-1-OL, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
4O9F
 
 | |
5B6E
 
 | |
3HX3
 
 | | Crystal structure of CRALBP mutant R234W | | Descriptor: | RETINAL, Retinaldehyde-binding protein 1 | | Authors: | Stocker, A, He, X, Lobsiger, J. | | Deposit date: | 2009-06-19 | | Release date: | 2009-10-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HY5
 
 | | Crystal structure of CRALBP | | Descriptor: | L(+)-TARTARIC ACID, RETINAL, Retinaldehyde-binding protein 1 | | Authors: | Stocker, A, He, X, Lobsiger, J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3P3Y
 
 | |
3R1V
 
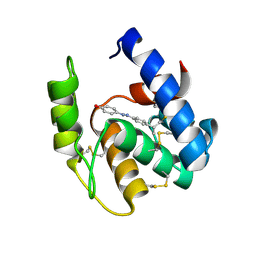 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges, in complex with an azo compound | | Descriptor: | 4-{(E)-[4-(propan-2-yl)phenyl]diazenyl}phenol, Odorant binding protein, antennal | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3R1P
 
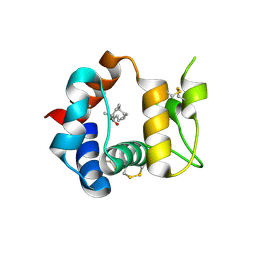 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges, form P1 | | Descriptor: | Odorant binding protein, antennal, PALMITIC ACID | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
2GFP
 
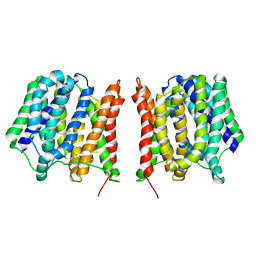 | | Structure of the Multidrug Transporter EmrD from Escherichia coli | | Descriptor: | Multidrug resistance protein D | | Authors: | Yin, Y, He, X, Szewczyk, P, Nguyen, T, Chang, G. | | Deposit date: | 2006-03-22 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the multidrug transporter EmrD from Escherichia coli
Science, 312, 2006
|
|
3R1O
 
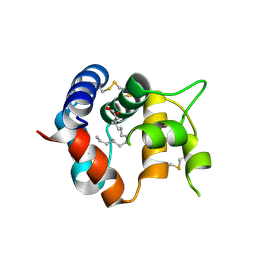 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges | | Descriptor: | Odorant binding protein, antennal, PALMITIC ACID | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3P40
 
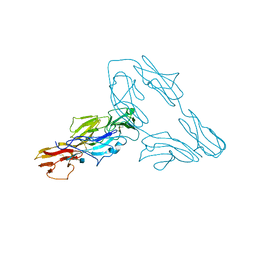 | |
5YYC
 
 | | Crystal structure of alanine racemase from Bacillus pseudofirmus (OF4) | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dong, H, Hu, T.T, He, G.Z, Lu, D.R, Qi, J.X, Dou, Y.S, Long, W, He, X, Su, D, Ju, J.S. | | Deposit date: | 2017-12-08 | | Release date: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural features and kinetic characterization of alanine racemase from Bacillus pseudofirmus OF4.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
4KGL
 
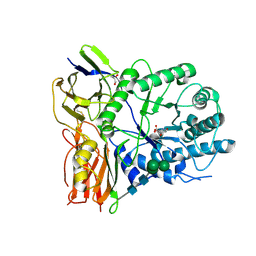 | | Crystal structure of human alpha-L-iduronidase complex with [2R,3R,4R,5S]-2-carboxy-3,4,5-trihydroxy-piperidine | | Descriptor: | (2R,3R,4R,5S)-3,4,5-trihydroxypiperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
4KGJ
 
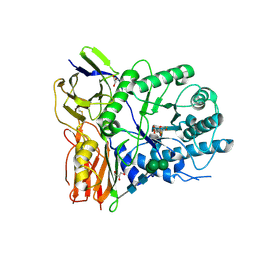 | | Crystal structure of human alpha-L-iduronidase complex with 5-fluoro-alpha-L-idopyranosyluronic acid fluoride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-fluoro-alpha-L-idopyranosyluronic acid fluoride, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
4KH2
 
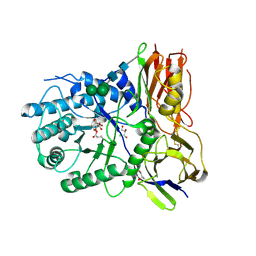 | | Crystal structure of human alpha-L-iduronidase complex with 2-deoxy-2-fluoro-alpha-L-idopyranosyluronic acid fluoride | | Descriptor: | 2,6-anhydro-5-deoxy-5-fluoro-L-idonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
8W87
 
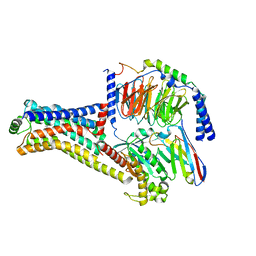 | | Cryo-EM structure of the METH-TAAR1 complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W8A
 
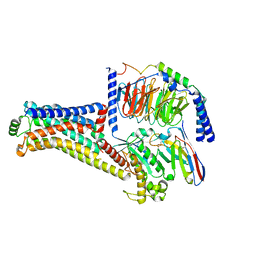 | | Cryo-EM structure of the RO5256390-TAAR1 complex | | Descriptor: | (4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-imine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W88
 
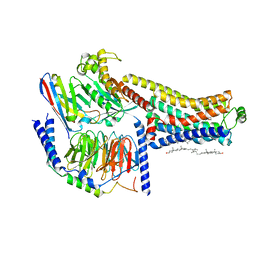 | | Cryo-EM structure of the SEP363856-bound TAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W8B
 
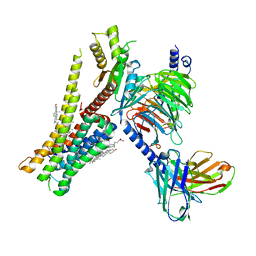 | | Cryo-EM structure of SEP-363856 bounded serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W89
 
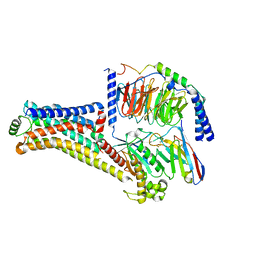 | | Cryo-EM structure of the PEA-bound TAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
7WP9
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPA
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPB
 
 | | SARS-CoV-2 Omicron Variant RBD complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
