7D8M
 
 | | Crystal structure of DyP | | Descriptor: | Dye-decolorizing peroxidase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | He, C, Jia, R, Wang, T, Li, L.Q. | | Deposit date: | 2020-10-08 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revealing two important tryptophan residues with completely different roles in a dye-decolorizing peroxidase from Irpex lacteus F17.
Biotechnol Biofuels, 14, 2021
|
|
4FZO
 
 | |
8HKD
 
 | |
6JJ8
 
 | | Crystal structure of OsHXK6-ATP-Mg2+ complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of OsHXK6-ATP-Mg2+ complex
To Be Published
|
|
6J2P
 
 | |
6J7M
 
 | |
1U8B
 
 | | Crystal structure of the methylated N-ADA/DNA complex | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*TP*T)-3', 5'-D(P*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*AP*T)-3', ... | | Authors: | He, C, Hus, J.-C, Sun, L.J, Zhou, P, Norman, D.P.G, Dotsch, V, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2004-08-05 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada.
Mol.Cell, 20, 2005
|
|
4HQE
 
 | | The crystal structure of QsrR-DNA complex | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*TP*AP*AP*TP*TP*AP*TP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*AP*TP*AP*AP*TP*TP*AP*TP*AP*CP*T)-3'), Transcriptional regulator QsrR | | Authors: | He, C, Ji, Q, Zhang, L. | | Deposit date: | 2012-10-25 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1ZGW
 
 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
3BTY
 
 | | Crystal structure of human ABH2 bound to dsDNA containing 1meA through cross-linking away from active site | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*DCP*DTP*DGP*DTP*DAP*DTP*(MA7)P*DAP*DCP*DTP*DGP*DCP*DG)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DGP*DTP*DTP*DAP*DTP*DAP*DCP*DA)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BTZ
 
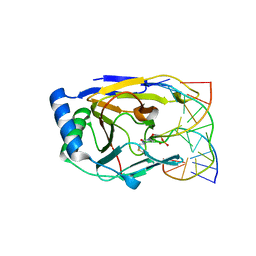 | | Crystal structure of human ABH2 cross-linked to dsDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*AP*GP*GP*TP*GP*AP*(2YR)P*AP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DTP*DTP*DAP*DTP*DCP*DAP*DCP*DC)-3') | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BUC
 
 | | X-ray structure of human ABH2 bound to dsDNA with Mn(II) and 2KG | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*DCP*DTP*DGP*DTP*DAP*DTP*(MA7)P*DAP*DCP*DTP*DGP*DCP*DG)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
4N4R
 
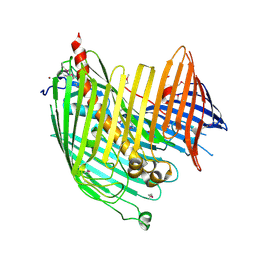 | | Structure basis of lipopolysaccharide biogenesis | | Descriptor: | CACODYLATE ION, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Dong, H, Xiang, Q, Wang, Z, Paterson, N.G, He, C, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2013-10-08 | | Release date: | 2014-06-25 | | Last modified: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for outer membrane lipopolysaccharide insertion.
Nature, 511, 2014
|
|
7WYG
 
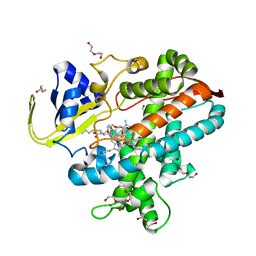 | | Crystal structure of P450BSbeta-L78I/Q85H/G290I variant in complex with palmitic acid. | | Descriptor: | Cytochrome P450 152A1, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Li, F, He, C, Wang, X. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biocatalytic Enantioselective beta-Hydroxylation of Unactivated C-H Bonds in Aliphatic Carboxylic Acids.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7RFP
 
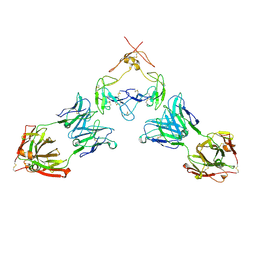 | | Mouse GITR (mGITR) with DTA-1 Fab fragment | | Descriptor: | DTA-1 (heavy chain), DTA-1 (light chain), Tumor necrosis factor receptor superfamily member 18,Enhanced green fluorescent protein | | Authors: | Meyerson, J.R, He, C. | | Deposit date: | 2021-07-14 | | Release date: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Therapeutic antibody activation of the glucocorticoid-induced TNF receptor by a clustering mechanism.
Sci Adv, 8, 2022
|
|
3MVF
 
 | |
7XPT
 
 | | Crystal structrue of MtdL:GDP:Mn soaked with GDP-Glc | | Descriptor: | GDP-ALPHA-D-GLUCOSE, MANGANESE (II) ION, Transglycosylse | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
7XPR
 
 | | Crystal structrue of SeMet-MtdL:GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transglycosylse | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
7XPV
 
 | | crysteal structure of MtdL-S228A-His soaked with GDP-Fucf and Mn | | Descriptor: | MANGANESE (II) ION, Transglycosylse, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanyl-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{S},3~{S},4~{S},5~{R})-3,4-bis(oxidanyl)-5-[(1~{S})-1-oxidanylethyl]oxolan-2-yl] hydrogen phosphate | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
7XPS
 
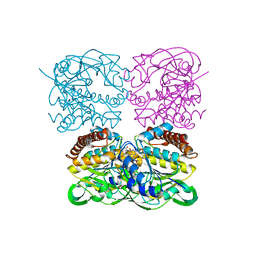 | | Crystal structrue of MtdL:GDP:Mn | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, Transglycosylse | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
7XPU
 
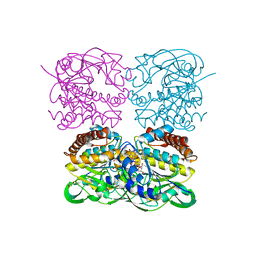 | | crystal structure of MtdL-S228A-His soaked GDP-Fucp and Mn | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, MANGANESE (II) ION, ... | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
4WQN
 
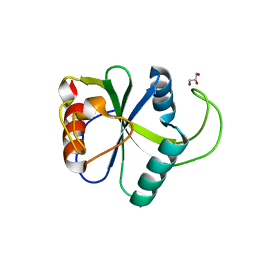 | | Crystal structure of N6-methyladenosine RNA reader YTHDF2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, YTH domain-containing family protein 2 | | Authors: | Zhu, T, Roundtree, I.A, Wang, P, Wang, X, Wang, L, Sun, C, Tian, Y, Li, J, He, C, Xu, Y. | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine.
Cell Res., 24, 2014
|
|
3R26
 
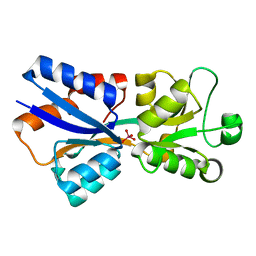 | | Perrhenate Binding to Molybdate Binding Protein | | Descriptor: | Molybdate-binding periplasmic protein, PERRHENATE | | Authors: | Aryal, B.P, Brugarolas, P, He, C. | | Deposit date: | 2011-03-13 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of ReO(4) (-) with an engineered MoO (4) (2-)-binding protein: towards a new approach in radiopharmaceutical applications.
J.Biol.Inorg.Chem., 17, 2012
|
|
4YWZ
 
 | |
5HQC
 
 | | A Glycoside Hydrolase Family 97 enzyme R171K variant from Pseudoalteromonas sp. strain K8 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
