3GE8
 
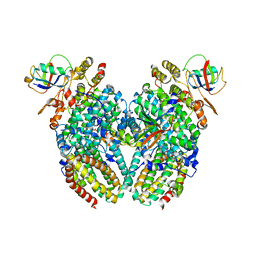 | | Toluene 4-monooxygenase HD T201A diferric, resting state complex | | Descriptor: | ACETATE ION, FE (III) ION, Toluene-4-monooxygenase system protein A, ... | | Authors: | Elsen, N.L, Bailey, L.J, Hauser, A.D, Fox, B.G. | | Deposit date: | 2009-02-25 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Role for threonine 201 in the catalytic cycle of the soluble diiron hydroxylase toluene 4-monooxygenase.
Biochemistry, 48, 2009
|
|
4NSB
 
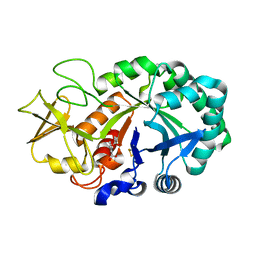 | | Crystal structure of the complex of signaling glycoprotein, SPB-40 and N-acetyl salicylic acid at 3.05 A resolution | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Yamini, S, Chaudhary, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the complex of signaling glycoprotein, SPB-40 and N-acetyl salicylic acid at 3.05 A resolution
To be Published
|
|
4AN7
 
 | |
6IYE
 
 | |
4CDN
 
 | | Crystal structure of M. mazei photolyase with its in vivo reconstituted 8-HDF antenna chromophore | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DEOXYRIBODIPYRIMIDINE PHOTOLYASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kiontke, S, Batschauer, A, Essen, L.-O. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Evolutionary Aspects of Antenna Chromophore Usage by Class II Photolyases.
J.Biol.Chem., 289, 2014
|
|
2XRY
 
 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | DEOXYRIBODIPYRIMIDINE PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
2XRZ
 
 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei in complex with intact CPD-lesion | | Descriptor: | ACETATE ION, COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
4ML4
 
 | | Crystal structure of the complex of signaling glycoprotein from buffalo (SPB-40) with tetrahydropyran at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, TETRAHYDROPYRAN | | Authors: | Yamini, S, Chaudhary, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of signaling glycoprotein from buffalo (SPB-40) with tetrahydropyran at 2.5 A resolution
To be Published
|
|
4OB9
 
 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution | | Descriptor: | Chorismate synthase | | Authors: | Shukla, P.K, Chaudhary, A, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution
To be Published
|
|
2WL7
 
 | | Crystal structure of the PSD93 PDZ1 domain | | Descriptor: | CHLORIDE ION, DISKS LARGE HOMOLOG 2, SULFATE ION | | Authors: | Fiorentini, M, Kallehauge, A, Kristensen, O, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2009-06-22 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.028 Å) | | Cite: | Structure of the First Pdz Domain of Human Psd-93.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2W84
 
 | | Structure of Pex14 in complex with Pex5 | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19.
EMBO J., 28, 2009
|
|
4MPK
 
 | | Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, GLYCEROL | | Authors: | Yamini, S, Chaudhary, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-13 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution
To be Published
|
|
3SNL
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 6-chloro-3,4-dimethyl-1-(3-methylpyridin-4-yl)-8-(trifluoromethyl)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
4MTV
 
 | | Crystal structure of the complex of Buffalo Signalling Glycoprotein with pentasaccharide at 2.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Shukla, P.K, Chaudhary, A, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the complex of Buffalo Signalling Glycoprotein with pentasaccharide at 2.8A resolution
To be Published
|
|
3SNI
 
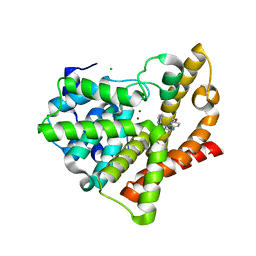 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 2-methoxy-6,7-dimethyl-9-(4-methylpyridin-3-yl)imidazo[1,5-a]pyrido[3,2-e]pyrazine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
1U8T
 
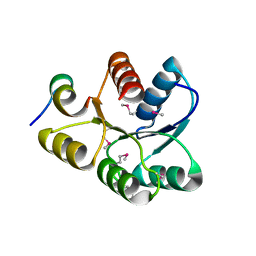 | | Crystal structure of CheY D13K Y106W alone and in complex with a FliM peptide | | Descriptor: | Chemotaxis protein cheY, Flagellar motor switch protein fliM, SULFATE ION | | Authors: | Dyer, C.M, Quillin, M.L, Campos, A, Lu, J, McEvoy, M.M, Hausrath, A.C, Westbrook, E.M, Matsumura, P, Matthews, B.W, Dahlquist, F.W. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Constitutively Active Double Mutant CheY(D13K Y106W) Alone and in Complex with a FliM Peptide
J.Mol.Biol., 342, 2004
|
|
4CDM
 
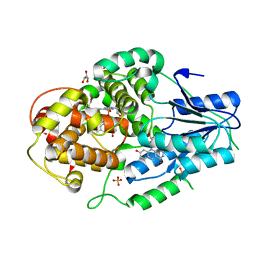 | | Crystal structure of M. mazei photolyase soaked with synthetic 8-HDF | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DEOXYRIBODIPYRIMIDINE PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kiontke, S, Batschauer, A, Essen, L.-O. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Evolutionary Aspects of Antenna Chromophore Usage by Class II Photolyases.
J.Biol.Chem., 289, 2014
|
|
4BK4
 
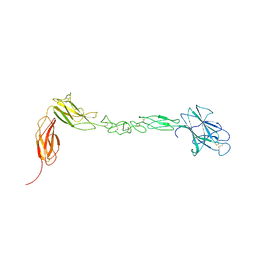 | | crystal structure of the human EphA4 ectodomain | | Descriptor: | EPHRIN TYPE-A RECEPTOR 4 | | Authors: | Seiradake, E, Schaupp, A, del Toro Ruiz, D, Kaufmann, R, Mitakidis, N, Harlos, K, Aricescu, A.R, Klein, R, Jones, E.Y. | | Deposit date: | 2013-04-22 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structurally Encoded Intraclass Differences in Epha Clusters Drive Distinct Cell Responses
Nat.Struct.Mol.Biol., 20, 2013
|
|
5Z9A
 
 | |
4BKA
 
 | | crystal structure of the human EphA4 ectodomain in complex with human ephrin A5 | | Descriptor: | EPHRIN TYPE-A RECEPTOR 4, EPHRIN-A5 | | Authors: | Seiradake, E, Schaupp, A, del Toro Ruiz, D, Kaufmann, R, Mitakidis, N, Harlos, K, Aricescu, A.R, Klein, R, Jones, E.Y. | | Deposit date: | 2013-04-23 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5.3 Å) | | Cite: | Structurally Encoded Intraclass Differences in Epha Clusters Drive Distinct Cell Responses
Nat.Struct.Mol.Biol., 20, 2013
|
|
8A5R
 
 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized and measured in dark. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EL222 from Erythrobacter litoralis.
To Be Published
|
|
8A5S
 
 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized in dark, measured illuminated. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EL222 from Erythrobacter litoralis.
To Be Published
|
|
4BK5
 
 | | crystal structure of the human EphA4 ectodomain in complex with human ephrin A5 (amine-methylated sample) | | Descriptor: | EPHRIN TYPE-A RECEPTOR 4, EPHRIN-A5 | | Authors: | Seiradake, E, Schaupp, A, del Toro Ruiz, D, Kaufmann, R, Mitakidis, N, Harlos, K, Aricescu, A.R, Klein, R, Jones, E.Y. | | Deposit date: | 2013-04-22 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structurally Encoded Intraclass Differences in Epha Clusters Drive Distinct Cell Responses
Nat.Struct.Mol.Biol., 20, 2013
|
|
4BKF
 
 | | crystal structure of the human EphA4 ectodomain in complex with human ephrinB3 | | Descriptor: | EPHRIN TYPE-A RECEPTOR 4, EPHRIN-B3 | | Authors: | Seiradake, E, Schaupp, A, del Toro Ruiz, D, Kaufmann, R, Mitakidis, N, Harlos, K, Aricescu, A.R, Klein, R, Jones, E.Y. | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.65 Å) | | Cite: | Structurally Encoded Intraclass Differences in Epha Clusters Drive Distinct Cell Responses
Nat.Struct.Mol.Biol., 20, 2013
|
|
2W85
 
 | | Structure of Pex14 in complex with Pex19 | | Descriptor: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
