6T6V
 
 | | Glu-494-Ala inactive monomer of a quinol dependent Nitric Oxide Reductase (qNOR) from Alcaligenes xylosoxidans | | Descriptor: | CALCIUM ION, Nitric oxide reductase subunit B, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gopalasingam, C.C, Johnson, R.M, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-10-19 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
6TFD
 
 | | Crystal structure of nitrite and NO bound three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
6TFO
 
 | | Crystal structure of as isolated three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
6SW6
 
 | |
6THE
 
 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
5OCF
 
 | | Crystal structure of nitric oxide bound to three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OCB
 
 | | Crystal structure of nitric oxide bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
6THF
 
 | | Crystal structure of two-domain Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
5ONY
 
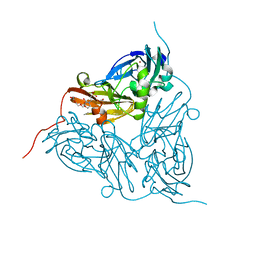 | | As-isolated resting state copper nitrite reductase from Achromobacter xylosoxidans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
5ONX
 
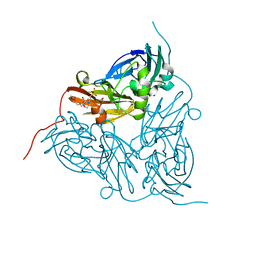 | | Resting state copper nitrite reductase determined by serial femtosecond rotation crystallography | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
5OKD
 
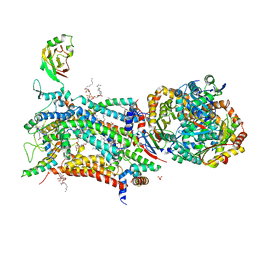 | | Crystal structure of bovine Cytochrome bc1 in complex with inhibitor SCR0911. | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, O'Neill, P.M, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2017-07-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
2PCR
 
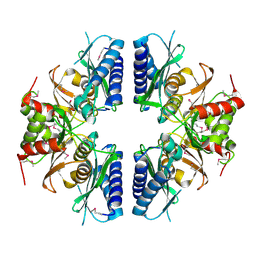 | | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5 | | Descriptor: | Inositol-1-monophosphatase | | Authors: | Jeyakanthan, J, Gayathri, D, Velmurugan, D, Agari, Y, Bessho, Y, Ellis, M.J, Antonyuk, S.V, Strange, R.W, Hasnain, S.S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5
To be Published
|
|
1NDT
 
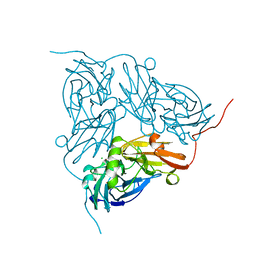 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (NITRITE REDUCTASE) | | Authors: | Dodd, F.E, Vanbeeumen, J, Eady, R.R, Hasnain, S.S. | | Deposit date: | 1998-10-28 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of a blue-copper nitrite reductase in two crystal forms. The nature of the copper sites, mode of substrate binding and recognition by redox partner.
J.Mol.Biol., 282, 1998
|
|
1NDS
 
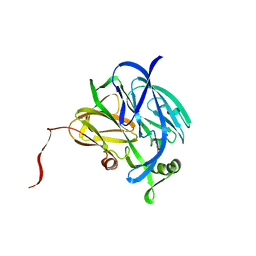 | | CRYSTALLOGRAPHIC STRUCTURE OF A SUBSTRATE BOUND BLUE COPPER NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | COPPER (II) ION, NITRITE ION, NITRITE REDUCTASE | | Authors: | Dodd, F.E, Hasnain, S.S, Abraham, Z.H.L, Eady, R.R, Smith, B.E. | | Deposit date: | 1997-01-23 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of a blue-copper nitrite reductase and its substrate-bound complex.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1NDR
 
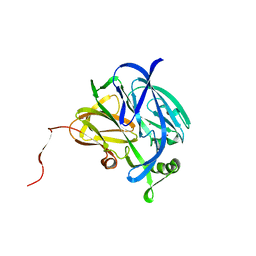 | | CRYSTALLOGRAPHIC STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Dodd, F.E, Hasnain, S.S, Abraham, Z.H.L, Eady, R.R, Smith, B.E. | | Deposit date: | 1997-01-23 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a blue-copper nitrite reductase and its substrate-bound complex.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1OZU
 
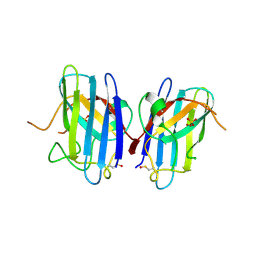 | | Crystal Structure of Familial ALS Mutant S134N of human Cu,Zn Superoxide Dismutase (CuZnSOD) to 1.3A resolution | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
3ZTZ
 
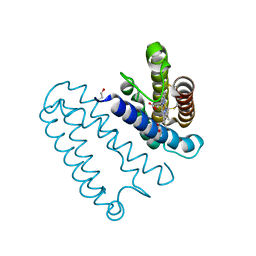 | | Cytochrome c prime from alcaligenes xylosoxidans: carbon monooxide bound L16G variant at 1.05 A resolution: unrestraint refinement | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-07-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1Q0E
 
 | |
3ZQV
 
 | |
4A7U
 
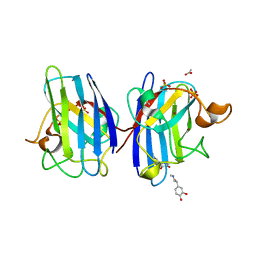 | | Structure of human I113T SOD1 complexed with adrenaline in the p21 space group. | | Descriptor: | ACETATE ION, COPPER (II) ION, L-EPINEPHRINE, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
4A7T
 
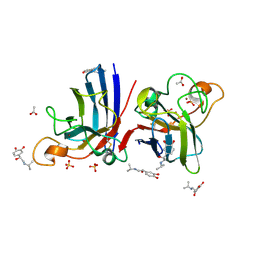 | | Structure of human I113T SOD1 mutant complexed with isoproteranol in the p21 space group | | Descriptor: | ACETATE ION, COPPER (II) ION, ISOPRENALINE, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
4A7Q
 
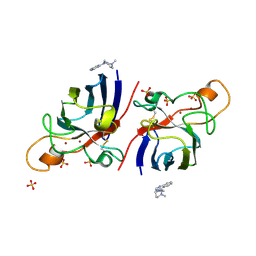 | | Structure of human I113T SOD1 mutant complexed with 4-(4-methyl-1,4- diazepan-1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYL-1,4-DIAZEPAN-1-YL)QUINAZOLINE, COPPER (II) ION, SULFATE ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-Ray Crystallography and Computational Docking for the Detection and Development of Protein-Ligand Interactions.
Curr.Med.Chem., 20, 2013
|
|
1OEZ
 
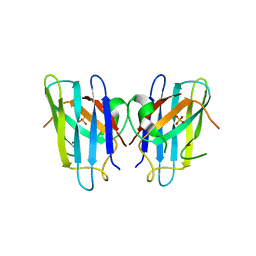 | | Zn His46Arg mutant of Human Cu, Zn Superoxide Dismutase | | Descriptor: | SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ZINC ION | | Authors: | Strange, R.W, Antonyuk, S, Hough, M.A, Doucette, P, Rodriguez, J, Elam, J.S, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2003-04-02 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Amyloid-Like Filaments and Water-Filled Nanotubes Formed by Sod1 Mutant Proteins Linked to Familial Als
Nat.Struct.Biol., 10, 2003
|
|
4A7S
 
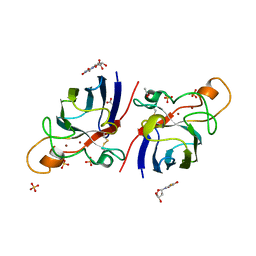 | | Structure of human I113T SOD1 mutant complexed with 5-Fluorouridine in the p21 space group | | Descriptor: | 5-FLUOROURIDINE, ACETATE ION, COPPER (II) ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-12-05 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
4A7G
 
 | | Structure of human I113T SOD1 mutant complexed with 4-methylpiperazin- 1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYLPIPERAZIN-1-YL)QUINAZOLINE, ACETATE ION, COPPER (II) ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Sharma, R, Antonyuk, S.V, Strange, R.W, Berry, N.G, O'Neil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | X-ray crystallography and computational docking for the detection and development of protein-ligand interactions.
Curr.Med.Chem., 20, 2013
|
|
