2TBV
 
 | |
3N0A
 
 | | Crystal structure of auxilin (40-400) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein phosphatase auxilin | | Authors: | Harrison, S.C, Guan, R, Dai, H, Kirchhausen, T. | | Deposit date: | 2010-05-13 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the PTEN-like Region of Auxilin, a Detector of Clathrin-Coated Vesicle Budding.
Structure, 18, 2010
|
|
1W63
 
 | | AP1 clathrin adaptor core | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 1 BETA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 SIGMA 1A SUBUNIT, ... | | Authors: | Heldwein, E, Macia, E, Wang, J, Yin, H.L, Kirchhausen, T, Harrison, S.C. | | Deposit date: | 2004-08-12 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of the Clathrin Adaptor Protein 1 Core
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8G0Q
 
 | |
5IBU
 
 | | 6652 Fab (unbound) | | Descriptor: | 6652 Heavy Chain, 6652 Light Chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain.
Nat. Med., 22, 2016
|
|
8G0P
 
 | |
6E4X
 
 | |
5W0D
 
 | |
5W6C
 
 | | UCA Fab (unbound) from 6649 Lineage | | Descriptor: | UCA Fab heavy chain, UCA Fab light chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2017-06-16 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4V7Q
 
 | | Atomic model of an infectious rotavirus particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Core scaffold protein, ... | | Authors: | Settembre, E.C, Chen, J.Z, Dormitzer, P.R, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2010-05-13 | | Release date: | 2014-07-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic model of an infectious rotavirus particle.
Embo J., 30, 2011
|
|
1MUK
 
 | | reovirus lambda3 native structure | | Descriptor: | MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
1MWH
 
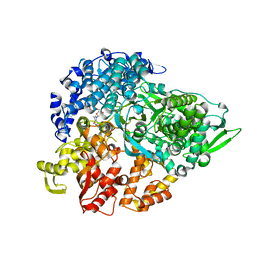 | | REOVIRUS POLYMERASE LAMBDA3 BOUND TO MRNA CAP ANALOG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MANGANESE (II) ION, MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-29 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
1N38
 
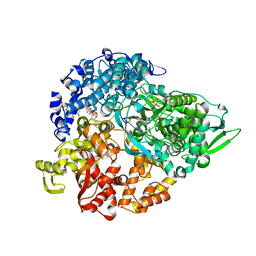 | | reovirus polymerase lambda3 elongation complex with one phosphodiester bond formed | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 3'-DEOXY-URIDINE 5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*C)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-25 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
1N35
 
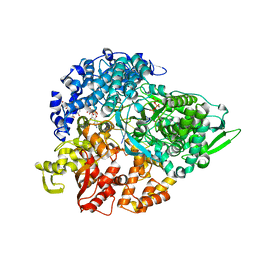 | | lambda3 elongation complex with four phosphodiester bond formed | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*CP*CP*CP*CP*C)-3', 5'-R(P*GP*GP*GP*GP*G)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-25 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
8V10
 
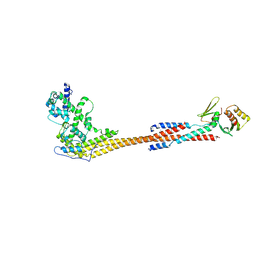 | |
8UK2
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 5 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
8V11
 
 | |
8UK3
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 6 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
5W94
 
 | |
7N64
 
 | | SARS-CoV-2 Spike (2P) in complex with G32R7 Fab (RBD and NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G32R7 Fab heavy chain, ... | | Authors: | Windsor, I.W, Jenni, S, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
7N62
 
 | | SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C12C9 Fab heavy chain, C12C9 Fab light chain, ... | | Authors: | Windsor, I.W, Jenni, S, Bajic, G, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
8T0P
 
 | | Structure of Cse4 bound to Ame1 and Okp1 | | Descriptor: | Histone H3-like centromeric protein CSE4, Inner kinetochore subunit AME1, Inner kinetochore subunit OKP1, ... | | Authors: | Deng, S, Harrison, S.C. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Recognition of centromere-specific histone Cse4 by the inner kinetochore Okp1-Ame1 complex.
Embo Rep., 24, 2023
|
|
7UMK
 
 | | Structure of vesicular stomatitis virus (helical reconstruction, 4.1 A resolution) | | Descriptor: | Matrix protein, Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Jenni, S, Horwitz, J.A, Bloyet, L.-M, Whelan, S.P.J, Harrison, S.C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle.
Nat Commun, 13, 2022
|
|
7UML
 
 | | Structure of vesicular stomatitis virus (local reconstruction, 3.5 A resolution) | | Descriptor: | Matrix protein, Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Jenni, S, Horwitz, J.A, Bloyet, L.-M, Whelan, S.P.J, Harrison, S.C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle.
Nat Commun, 13, 2022
|
|
8UDG
 
 | | S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004 | | Descriptor: | Hemagglutinin, S1V2-72 heavy chain, S1V2-72 light chain | | Authors: | Finney, J, Kong, S, Walsh Jr, R.M, Harrison, S.C, Kelsoe, G. | | Deposit date: | 2023-09-28 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (4.98 Å) | | Cite: | Protective human antibodies against a conserved epitope in pre- and postfusion influenza hemagglutinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
