3CRO
 
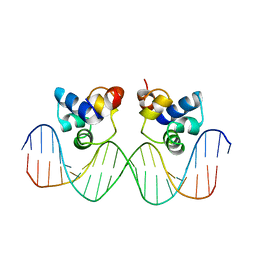 | | THE PHAGE 434 CRO/OR1 COMPLEX AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*AP*CP*AP*AP*AP*CP*TP*TP*TP*CP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*GP*AP*AP*AP*GP*TP*TP*TP*GP*T P*AP*CP*T)-3'), PROTEIN (434 CRO) | | Authors: | Mondragon, A, Harrison, S.C. | | Deposit date: | 1990-07-06 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The phage 434 Cro/OR1 complex at 2.5 A resolution.
J.Mol.Biol., 219, 1991
|
|
5IBT
 
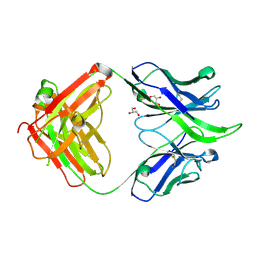 | | UCA Fab (unbound) from 6515 Lineage | | Descriptor: | GLYCEROL, UCA Heavy Chain, UCA Light Chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain.
Nat. Med., 22, 2016
|
|
5IBL
 
 | |
5IBU
 
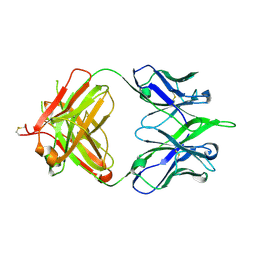 | | 6652 Fab (unbound) | | Descriptor: | 6652 Heavy Chain, 6652 Light Chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain.
Nat. Med., 22, 2016
|
|
4IT3
 
 | | Crystal Structure of Iml3 from S. cerevisiae | | Descriptor: | Central kinetochore subunit IML3 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2013-01-17 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | An iml3-chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation.
Cell Rep, 5, 2013
|
|
4JE3
 
 | |
6U1X
 
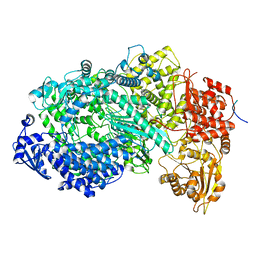 | | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor (3.0 A resolution) | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Jenni, S, Bloyet, L.M, Dias-Avalos, R, Liang, B, Wheelman, S.P.J, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2019-08-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor.
Cell Rep, 30, 2020
|
|
6UEB
 
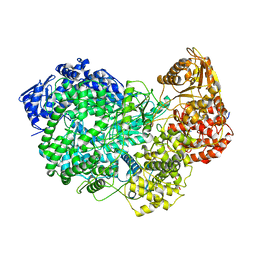 | |
6E8W
 
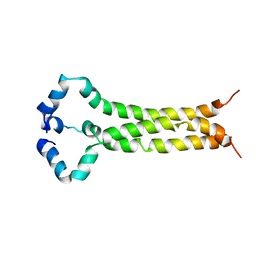 | | MPER-TM Domain of HIV-1 envelope glycoprotein (Env) | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Fu, Q, Shaik, M.M, Cai, Y, Ghantous, F, Piai, A, Peng, H, Rits-Volloch, S, Liu, Z, Harrison, S.C, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane proximal external region of HIV-1 envelope glycoprotein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7N64
 
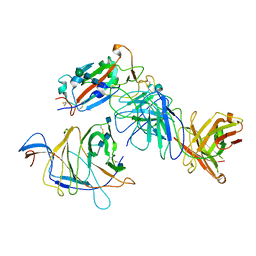 | | SARS-CoV-2 Spike (2P) in complex with G32R7 Fab (RBD and NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G32R7 Fab heavy chain, ... | | Authors: | Windsor, I.W, Jenni, S, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
7N62
 
 | | SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C12C9 Fab heavy chain, C12C9 Fab light chain, ... | | Authors: | Windsor, I.W, Jenni, S, Bajic, G, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
5T6J
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | Kinetochore protein SPC24, Kinetochore protein SPC25, Kinetochore-associated protein DSN1 | | Authors: | Valverde, R, Jenni, S, Dimitrova, Y, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
6XPX
 
 | |
6XPO
 
 | | Influenza hemagglutinin A/Bangkok/01/1979(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | McCarthy, K.R, Harrison, S.C. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Prevalent Focused Human Antibody Response to the Influenza Virus Hemagglutinin Head Interface.
Mbio, 12, 2021
|
|
5T51
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | KLLA0E05809p, KLLA0F02343p, SULFATE ION | | Authors: | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2007 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
5T59
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, KLLA0B13629p, KLLA0E05809p, ... | | Authors: | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
6E56
 
 | |
7L7Q
 
 | | Ctf3c with Ulp2-KIM | | Descriptor: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2020-12-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ctf3/CENP-I provides a docking site for the desumoylase Ulp2 at the kinetochore.
J.Cell Biol., 220, 2021
|
|
7KDF
 
 | | Structure of Stu2 Bound to dwarf Ndc80c | | Descriptor: | NDC80 isoform 1,NDC80 isoform 1, NUF2 isoform 1,NUF2 isoform 1, SPC25 isoform 1,SPC25 isoform 1, ... | | Authors: | Zahm, J.A, Stewart, M.G, Miller, M.P, Harrison, S.C. | | Deposit date: | 2020-10-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of Stu2 recruitment to yeast kinetochores.
Elife, 10, 2021
|
|
5T58
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | KLLA0C15939p, KLLA0D15741p, KLLA0E05809p, ... | | Authors: | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2131 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
6V05
 
 | | Cryo-EM structure of a substrate-engaged Bam complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Tomasek, D, Rawson, S, Lee, J, Wzorek, J.S, Harrison, S.C, Li, Z, Kahne, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a nascent membrane protein as it folds on the BAM complex.
Nature, 583, 2020
|
|
5A22
 
 | | Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy | | Descriptor: | VESICULAR STOMATITIS VIRUS L POLYMERASE, ZINC ION | | Authors: | Liang, B, Li, Z, Jenni, S, Rameh, A.A, Morin, B.M, Grant, T, Grigorieff, N, Harrison, S.C, Whelan, S.P.J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-08-19 | | Last modified: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy.
Cell(Cambridge,Mass.), 162, 2015
|
|
6ULC
 
 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab at a nominal resolution of 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Chen, B, Harrison, S.C. | | Deposit date: | 2019-10-07 | | Release date: | 2020-04-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
5TD8
 
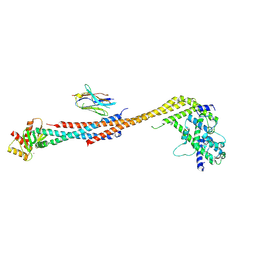 | | Crystal structure of an Extended Dwarf Ndc80 Complex | | Descriptor: | Kinetochore protein NDC80, Kinetochore protein NUF2, Kinetochore protein SPC24, ... | | Authors: | Valverde, R, Ingram, J, Harrison, S.C. | | Deposit date: | 2016-09-17 | | Release date: | 2016-11-16 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (7.531 Å) | | Cite: | Conserved Tetramer Junction in the Kinetochore Ndc80 Complex.
Cell Rep, 17, 2016
|
|
6WXF
 
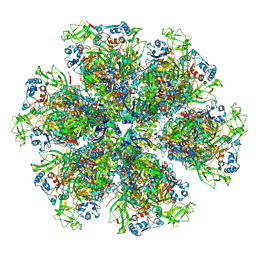 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Intermediate conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
