2VPV
 
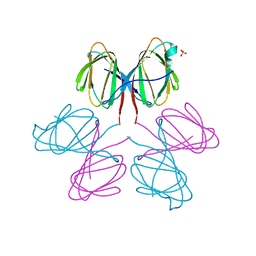 | | Dimerization Domain of Mif2p | | Descriptor: | PROTEIN MIF2, SULFATE ION | | Authors: | Cohen, R.L, Espelin, C.W, Sorger, P.K, Harrison, S.C, Simons, K.T. | | Deposit date: | 2008-03-05 | | Release date: | 2008-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Dissection of Mif2P, a Conserved DNA-Binding Kinetochore Protein.
Mol.Biol.Cell, 19, 2008
|
|
7UI0
 
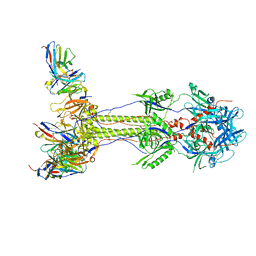 | | Post-fusion ectodomain of HSV-1 gB in complex with HSV010-13 Fab | | Descriptor: | Envelope glycoprotein B, HSV10-13 Fab Heavy chain, HSV10-13 Light chain | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
7UHZ
 
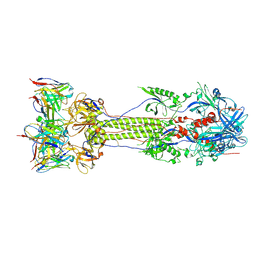 | | Post-fusion ectodomain of HSV-1 gB in complex with BMPC-23 Fab | | Descriptor: | BMPC-23 Fab Heavy chain, BMPC-23 Fab Light chain, Envelope glycoprotein B | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
1FAV
 
 | | THE STRUCTURE OF AN HIV-1 SPECIFIC CELL ENTRY INHIBITOR IN COMPLEX WITH THE HIV-1 GP41 TRIMERIC CORE | | Descriptor: | HIV-1 ENVELOPE PROTEIN CHIMERA, PROTEIN (TRANSMEMBRANE GLYCOPROTEIN) | | Authors: | Zhou, G, Ferrer, M, Chopra, R, Strassmaier, T, Weissenhorn, W, Skehel, J.J, Oprian, D, Schreiber, S.L, Harrison, S.C, Wiley, D.C. | | Deposit date: | 2000-07-13 | | Release date: | 2000-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of an HIV-1 specific cell entry inhibitor in complex with the HIV-1 gp41 trimeric core.
Bioorg.Med.Chem., 8, 2000
|
|
6WXF
 
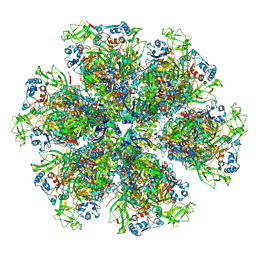 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Intermediate conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
2YFW
 
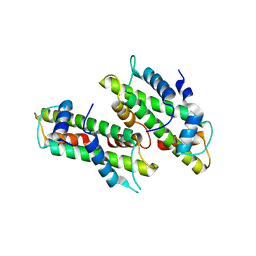 | |
6WXG
 
 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Reversed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
3IYV
 
 | | Clathrin D6 coat as full-length Triskelions | | Descriptor: | Clathrin heavy chain, Clathrin light chain A | | Authors: | Johnson, G.T, Fotin, A, Cheng, Y, Sliz, P, Grigorieff, N, Harrison, S.C, Kirchhausen, T, Walz, T. | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular model for a complete clathrin lattice from electron cryomicroscopy.
Nature, 432, 2004
|
|
1C9L
 
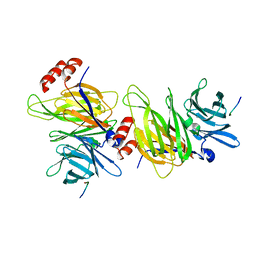 | |
1C9I
 
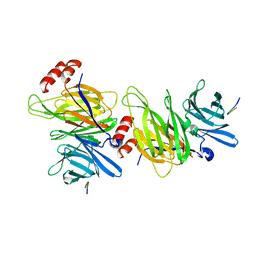 | |
6U1X
 
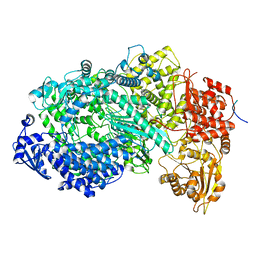 | | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor (3.0 A resolution) | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Jenni, S, Bloyet, L.M, Dias-Avalos, R, Liang, B, Wheelman, S.P.J, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2019-08-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor.
Cell Rep, 30, 2020
|
|
6OUA
 
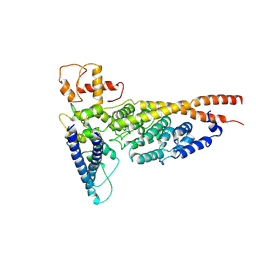 | | Cryo-EM structure of the yeast Ctf3 complex | | Descriptor: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2019-05-04 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | The structure of the yeast Ctf3 complex.
Elife, 8, 2019
|
|
1ZXQ
 
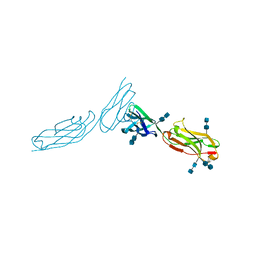 | | THE CRYSTAL STRUCTURE OF ICAM-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-2 | | Authors: | Casasnovas, J.M, Springer, T.A, Harrison, S.C, Wang, J.-H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ICAM-2 reveals a distinctive integrin recognition surface.
Nature, 387, 1997
|
|
4XDN
 
 | | Crystal structure of Scc4 in complex with Scc2n | | Descriptor: | MAU2 chromatid cohesion factor homolog, SULFATE ION, Sister chromatid cohesion protein 2 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2014-12-19 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural evidence for Scc4-dependent localization of cohesin loading.
Elife, 4, 2015
|
|
1FN9
 
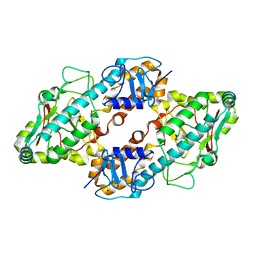 | | CRYSTAL STRUCTURE OF THE REOVIRUS OUTER CAPSID PROTEIN SIGMA 3 | | Descriptor: | OUTER-CAPSID PROTEIN SIGMA 3, ZINC ION | | Authors: | Olland, A.M, Jane-Valbuena, J, Schiff, L.A, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2000-08-21 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the reovirus outer capsid and dsRNA-binding protein sigma3 at 1.8 A resolution.
EMBO J., 20, 2001
|
|
1YMH
 
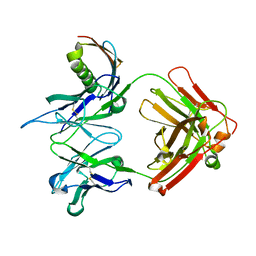 | | anti-HCV Fab 19D9D6 complexed with protein L (PpL) mutant A66W | | Descriptor: | Fab 16D9D6, heavy chain, light chain, ... | | Authors: | Granata, V, Housden, N.G, Harrison, S, Jolivet-Reynaud, C, Gore, M.G, Stura, E.A. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of the crystallization and crystal packing of two Fab single-site mutant protein L complexes.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6V05
 
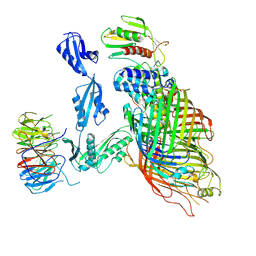 | | Cryo-EM structure of a substrate-engaged Bam complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Tomasek, D, Rawson, S, Lee, J, Wzorek, J.S, Harrison, S.C, Li, Z, Kahne, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a nascent membrane protein as it folds on the BAM complex.
Nature, 583, 2020
|
|
1N38
 
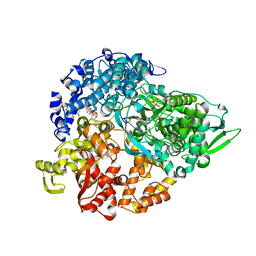 | | reovirus polymerase lambda3 elongation complex with one phosphodiester bond formed | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 3'-DEOXY-URIDINE 5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*C)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-25 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
1MWH
 
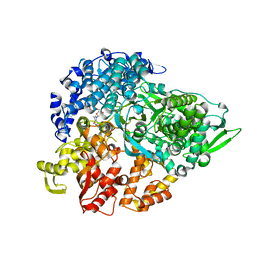 | | REOVIRUS POLYMERASE LAMBDA3 BOUND TO MRNA CAP ANALOG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MANGANESE (II) ION, MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-29 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
1MUK
 
 | | reovirus lambda3 native structure | | Descriptor: | MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
1N35
 
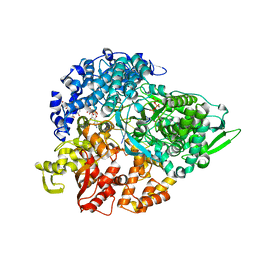 | | lambda3 elongation complex with four phosphodiester bond formed | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*CP*CP*CP*CP*C)-3', 5'-R(P*GP*GP*GP*GP*G)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-25 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
1N1H
 
 | | Initiation complex of polymerase lambda3 from reovirus | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*C)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-17 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
8SMV
 
 | | GPR161 Gs heterotrimer | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hoppe, N, Manglik, A, Harrison, S. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1NFI
 
 | | I-KAPPA-B-ALPHA/NF-KAPPA-B COMPLEX | | Descriptor: | I-KAPPA-B-ALPHA, NF-KAPPA-B P50, NF-KAPPA-B P65 | | Authors: | Jacobs, M.D, Harrison, S.C. | | Deposit date: | 1998-08-25 | | Release date: | 1998-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an IkappaBalpha/NF-kappaB complex.
Cell(Cambridge,Mass.), 95, 1998
|
|
6PPE
 
 | | ClpP and ClpX IGF loop in ClpX-ClpP complex with D7 symmetry | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
