6OUA
 
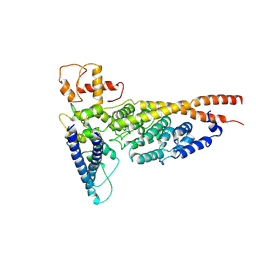 | | Cryo-EM structure of the yeast Ctf3 complex | | Descriptor: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2019-05-04 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | The structure of the yeast Ctf3 complex.
Elife, 8, 2019
|
|
5IBL
 
 | |
2P6B
 
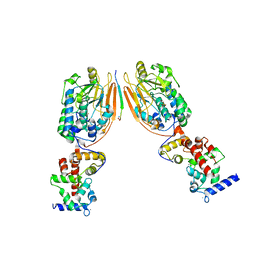 | | Crystal Structure of Human Calcineurin in Complex with PVIVIT Peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B isoform 1, Calmodulin-dependent calcineurin A subunit alpha isoform, ... | | Authors: | Li, H, Zhang, L, Rao, A, Harrison, S.C, Hogan, P.G. | | Deposit date: | 2007-03-16 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of calcineurin in complex with PVIVIT peptide: Portrait of a low-affinity signalling interaction
J.Mol.Biol., 369, 2007
|
|
1KRI
 
 | |
1KQR
 
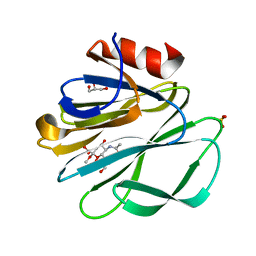 | | Crystal Structure of the Rhesus Rotavirus VP4 Sialic Acid Binding Domain in Complex with 2-O-methyl-alpha-D-N-acetyl neuraminic acid | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Dormitzer, P.R, Sun, Z.-Y.J, Wagner, G, Harrison, S.C. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Rhesus Rotavirus VP4 Sialic Acid Binding Domain has a Galectin Fold with a
Novel Carbohydrate Binding Site
Embo J., 21, 2002
|
|
3ZX8
 
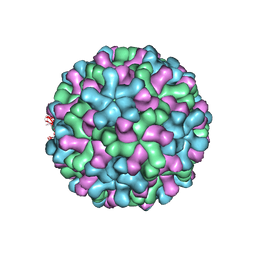 | | Cryo-EM reconstruction of native and expanded Turnip Crinkle virus | | Descriptor: | CAPSID PROTEIN | | Authors: | Bakker, S.E, Robottom, J, Hogle, J.M, Maeda, A, Pearson, A.R, Stockley, P.G, Ranson, N.A, Harrison, S.C. | | Deposit date: | 2011-08-08 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Isolation of an Asymmetric RNA Uncoating Intermediate for a Single-Stranded RNA Plant Virus.
J.Mol.Biol., 417, 2012
|
|
3ZXA
 
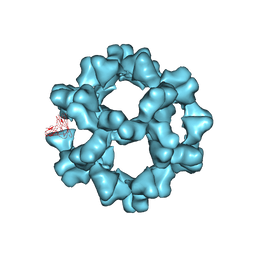 | |
6E4X
 
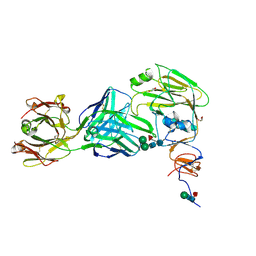 | |
6E56
 
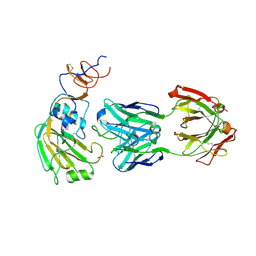 | |
3ZXU
 
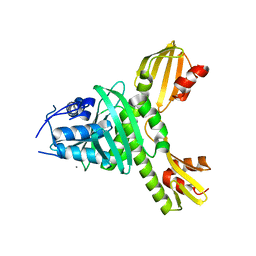 | |
8SMV
 
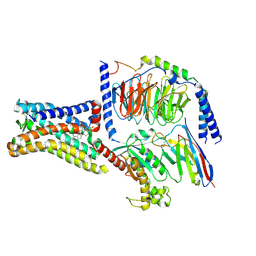 | | GPR161 Gs heterotrimer | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hoppe, N, Manglik, A, Harrison, S. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Biorxiv, 2023
|
|
8UDG
 
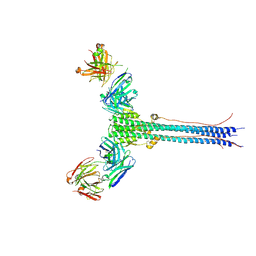 | | S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004 | | Descriptor: | Hemagglutinin, S1V2-72 heavy chain, S1V2-72 light chain | | Authors: | Finney, J, Kong, S, Walsh Jr, R.M, Harrison, S.C, Kelsoe, G. | | Deposit date: | 2023-09-28 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (4.98 Å) | | Cite: | Protective human antibodies against a conserved epitope in pre- and postfusion influenza hemagglutinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8G0Q
 
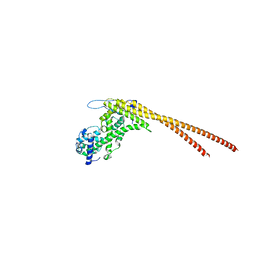 | |
8G0P
 
 | |
8UK2
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 5 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
8UK3
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 6 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
5KTB
 
 | |
8T0P
 
 | | Structure of Cse4 bound to Ame1 and Okp1 | | Descriptor: | Histone H3-like centromeric protein CSE4, Inner kinetochore subunit AME1, Inner kinetochore subunit OKP1, ... | | Authors: | Deng, S, Harrison, S.C. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Recognition of centromere-specific histone Cse4 by the inner kinetochore Okp1-Ame1 complex.
Embo Rep., 24, 2023
|
|
8V11
 
 | |
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
8V10
 
 | |
6E8W
 
 | | MPER-TM Domain of HIV-1 envelope glycoprotein (Env) | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Fu, Q, Shaik, M.M, Cai, Y, Ghantous, F, Piai, A, Peng, H, Rits-Volloch, S, Liu, Z, Harrison, S.C, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane proximal external region of HIV-1 envelope glycoprotein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6XPX
 
 | |
6XPO
 
 | | Influenza hemagglutinin A/Bangkok/01/1979(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | McCarthy, K.R, Harrison, S.C. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Prevalent Focused Human Antibody Response to the Influenza Virus Hemagglutinin Head Interface.
Mbio, 12, 2021
|
|
