3D6C
 
 | | Crystal structure of Staphylococcal nuclease variant PHS L38E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Harms, M.J, Schlessman, J.L, Sue, G.R, Garcia-Moreno, E.B. | | Deposit date: | 2008-05-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pK(a) values of acidic and basic residues buried at the same internal location in a protein are governed by different factors.
J.Mol.Biol., 389, 2009
|
|
3D4W
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS A109R at cryogenic temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Thermonuclease | | Authors: | Harms, M.J, Schlessman, J.L, Garcia-Moreno, E.B. | | Deposit date: | 2008-05-15 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arginine residues at internal positions in a protein are always charged.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3DHQ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS A90R at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Harms, M.J, Schlessman, J.L, Garcia-Moreno, E.B. | | Deposit date: | 2008-06-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Arginine residues at internal positions in a protein are always charged.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3D8G
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I72R at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Harms, M.J, Schlessman, J.L, Garcia-Moreno, E.B. | | Deposit date: | 2008-05-23 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Arginine residues at internal positions in a protein are always charged.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4FNE
 
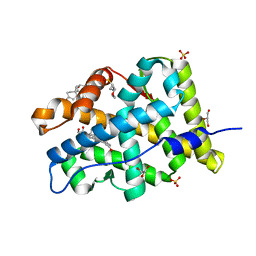 | | X-ray Crystal structure of the Ancestral 3-keto steroid receptor - DOC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DESOXYCORTICOSTERONE, ... | | Authors: | Ortlund, E.A, Colucci, J.K, Thornton, J, Eick, G, Harms, M. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Evolution of minimal specificity and promiscuity in steroid hormone receptors.
PLoS Genet, 8, 2012
|
|
6WN7
 
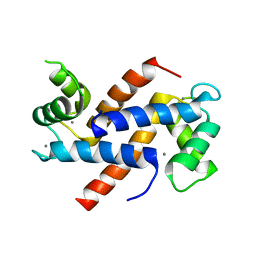 | | Homo sapiens S100A5 | | Descriptor: | CALCIUM ION, Protein S100-A5 | | Authors: | Perkins, A, Harms, M.J, Wong, C.E, Wheeler, L.C. | | Deposit date: | 2020-04-22 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Learning peptide recognition rules for a low-specificity protein.
Protein Sci., 29, 2020
|
|
7UI5
 
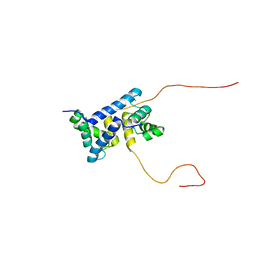 | | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9 | | Descriptor: | CALCIUM ION, Protein S100-A9 | | Authors: | Reardon, P.N, Harman, J.L, Costello, S.M, Warren, G.D, Phillips, S.R, Connor, P.J, Marqusee, S, Harms, M.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2RKS
 
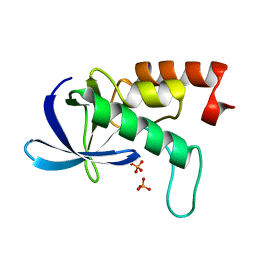 | |
4LY7
 
 | | Ancestral RNase H | | Descriptor: | Ribonuclease H, SULFATE ION | | Authors: | Hart, K.M, Harms, M.J, Schmidt, B.H, Elya, C, Thornton, J.W, Marqusee, S. | | Deposit date: | 2013-07-30 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Thermodynamic system drift in protein evolution.
Plos Biol., 12, 2014
|
|
8CGX
 
 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using XDS | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CGY
 
 | | Trypanosoma brucei IMP dehydrogenase (ori) crystallized in High Five cells reveals native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo; XDS processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD5
 
 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C53
 
 | | Trypanosoma brucei IMP dehydrogenase (ori) crystallized in High Five cells reveals native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo; CrystFEL processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C5K
 
 | | HEX-1 (in cellulo, in situ) crystallized and diffracted in High Five cells. Growth and SX data collection at 296 K on CrystalDirect plates | | Descriptor: | Woronin body major protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C51
 
 | | Trypanosoma brucei IMP dehydrogenase (cyto) crystallized in High Five cells revealing native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD4
 
 | | structure of HEX-1 from N. crassa crystallized in cellulo (cytosol), diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD6
 
 | | structure of HEX-1 (cyto V2) from N. crassa grown in living insect cells, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
1ZF9
 
 | | GGG Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*CP*GP*GP*GP*GP*G)-3', SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZFM
 
 | | AGC Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*CP*TP*AP*GP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZEX
 
 | | CCG A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*GP*GP*CP*CP*GP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZF2
 
 | | Four-stranded DNA Holliday Junction (CCC) | | Descriptor: | 5'-D(*CP*CP*GP*GP*GP*CP*CP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZFC
 
 | | ATC Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*AP*TP*AP*TP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZF4
 
 | | ATC Four-stranded DNA Holliday Junction | | Descriptor: | 5'-D(*CP*CP*GP*AP*TP*AP*TP*CP*GP*G)-3', SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZFF
 
 | | TTC Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*TP*TP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZEY
 
 | | CGG A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*GP*CP*GP*GP*GP*G)-3', SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
