7ZJ2
 
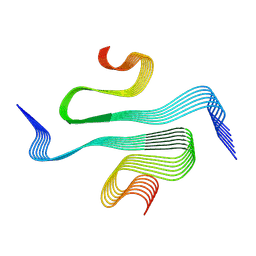 | |
8QXB
 
 | | TDP-43 amyloid fibrils: Morphology-2 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QXA
 
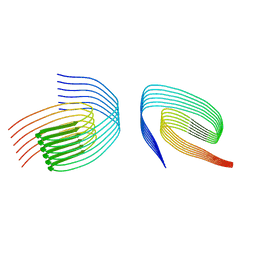 | | TDP-43 amyloid fibrils: Morphology-1b | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QX9
 
 | | TDP-43 amyloid fibrils: Morphology-1a | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
1P0L
 
 | | HP (2-20) Substitution GLN To TRP Modification In SDS-D25 MICELLES | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
1P5L
 
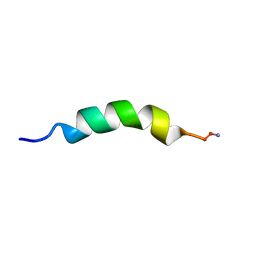 | | HP (2-20) Substitution PHE5 to SER modification in sds-d25 micelles | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-27 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
1P0J
 
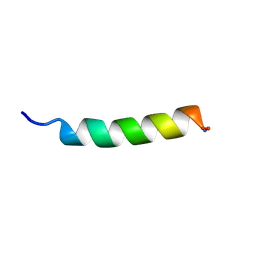 | | HP (2-20) Substitution ASP To TRP Modification In SDS-D25 Micelles | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
1P5K
 
 | | HP (2-20) Substitution SER to LEU11 modification in sds-d25 micelles | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-27 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
1P0G
 
 | | Structure of Antimicrobial Peptide, HP (2-20) and its Analogues Derived from Helicobacter pylori, as Determined by 1H NMR Spectroscopy | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
1P0O
 
 | | HP (2-20) substitution of Trp for Gln and Asp at position 17 and 19 MODIFICATION IN SDS-D25 MICELLES | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
5JQF
 
 | | Crystal structure of the lasso peptide Sphingopyxin I (SpI) | | Descriptor: | Sphingopyxin I | | Authors: | Fage, C.D, Hegemann, J.D, Harms, K, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-04 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
2YAK
 
 | | Structure of death-associated protein Kinase 1 (dapk1) in complex with a ruthenium octasporine ligand (OSV) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 1, RUTHENIUM OCTASPORINE 4 | | Authors: | Feng, L, Geisselbrecht, Y, Blanck, S, Wilbuer, A, Atilla-Gokcumen, G.E, Filippakopoulos, P, Kraeling, K, Celik, M.A, Harms, K, Maksimoska, J, Marmorstein, R, Frenking, G, Knapp, S, Essen, L.-O, Meggers, E. | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structurally Sophisticated Octahedral Metal Complexes as Highly Selective Protein Kinase Inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
2MFV
 
 | |
5J50
 
 | | Structure of tetrameric jacalin complexed with Gal beta-(1,3) GalNAc-alpha-OPNP | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of tetrameric jacalin complexed with Gal beta-(1,3) GalNAc-alpha-OPNP
To Be Published
|
|
5J4T
 
 | | Structure of tetrameric jacalin complexed with GlcNAc beta-(1,3) Gal-beta-OMe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Distortion of the ligand molecule as a strategy for modulating binding affinity: Further studies involving complexes of jacalin with beta-substituted disaccharides.
IUBMB Life, 69, 2017
|
|
5J4X
 
 | | Structure of tetrameric jacalin complexed with Gal beta-(1,3) Gal-beta-OMe | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Distortion of the ligand molecule as a strategy for modulating binding affinity: Further studies involving complexes of jacalin with beta-substituted disaccharides.
IUBMB Life, 69, 2017
|
|
4AS0
 
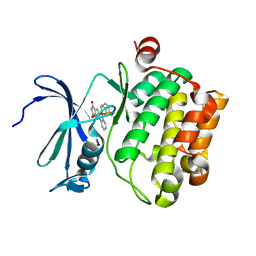 | | Cyclometalated Phthalimides as Protein Kinase Inhibitors | | Descriptor: | PHTALIMIDE-RUTHENIUM COMPLEX, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Blanck, S, Geisselbrecht, Y, Middel, S, Mietke, T, Harms, K, Essen, L.-O, Meggers, E. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bioactive Cyclometalated Phthalimides: Design, Synthesis and Kinase Inhibition.
Dalton Trans, 41, 2012
|
|
5D9E
 
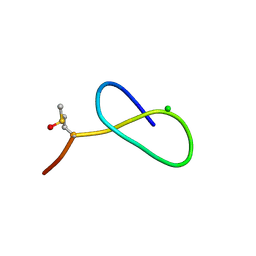 | | Crystal Structure of the Proline-rich Lasso Peptide Caulosegnin II | | Descriptor: | CHLORIDE ION, Caulosegnin II | | Authors: | Fage, C.D, Hegemann, J.D, Harms, K, Marahiel, M.A. | | Deposit date: | 2015-08-18 | | Release date: | 2016-02-17 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (0.859 Å) | | Cite: | The ring residue proline 8 is crucial for the thermal stability of the lasso peptide caulosegnin II.
Mol Biosyst, 12, 2016
|
|
4R6O
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NAG
 
 | | Xanthomonins I III are a New Class of Lasso Peptides Featuringa Seven-Membered Macrolactam Ring | | Descriptor: | HEXANE-1,6-DIOL, Xanthomonin I | | Authors: | Hegemann, J.D, Zimmermann, M, Zhu, S, Steuber, H, Harms, K, Xie, X, Marahiel, M.A. | | Deposit date: | 2013-10-22 | | Release date: | 2014-04-30 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Xanthomonins I-III: A New Class of Lasso Peptides with a Seven-Residue Macrolactam Ring.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
