2CY1
 
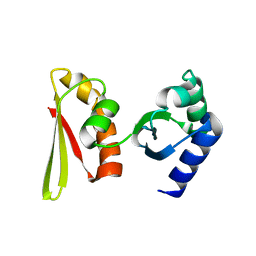 | | Crystal structure of APE1850 | | Descriptor: | NusA protein homolog | | Authors: | Shibata, R, Bessho, Y, Umehara, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of the archaeal transcription termination factor NusA: a significant decrease in twinning under microgravity conditions
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1WRU
 
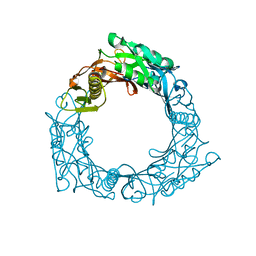 | | Structure of central hub elucidated by X-ray analysis of gene product 44; baseplate component of bacteriophage Mu | | Descriptor: | 43 kDa tail protein | | Authors: | Kondou, Y, Kitazawa, D, Takeda, S, Yamashita, E, Mizuguchi, M, Kawano, K, Tsukihara, T. | | Deposit date: | 2004-10-27 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of central hub elucidated by X-ray analysis of gene product 44; baseplate component of bacteriophage Mu
To be Published
|
|
2D1O
 
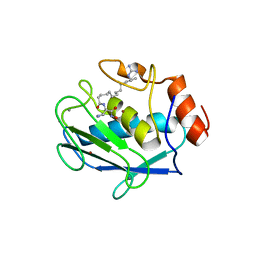 | | Stromelysin-1 (MMP-3) complexed to a hydroxamic acid inhibitor | | Descriptor: | CALCIUM ION, SM-25453, Stromelysin-1, ... | | Authors: | Kohno, T, Hochigai, H, Yamashita, E, Tsukihara, T, Kanaoka, M. | | Deposit date: | 2005-08-30 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of the catalytic domain of human stromelysin-1 (MMP-3) and collagenase-3 (MMP-13) with a hydroxamic acid inhibitor SM-25453
Biochem.Biophys.Res.Commun., 344, 2006
|
|
1IMJ
 
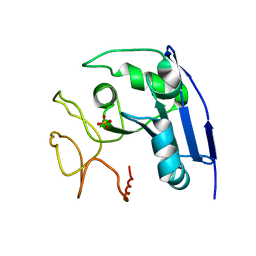 | |
2D1N
 
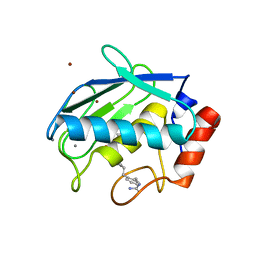 | | Collagenase-3 (MMP-13) complexed to a hydroxamic acid inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, SM-25453, ... | | Authors: | Kohno, T, Hochigai, H, Yamashita, E, Tsukihara, T, Kanaoka, M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structures of the catalytic domain of human stromelysin-1 (MMP-3) and collagenase-3 (MMP-13) with a hydroxamic acid inhibitor SM-25453
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2ROZ
 
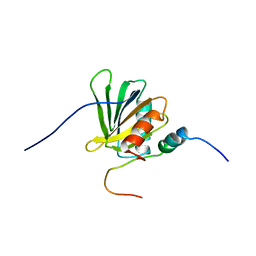 | | Structure of the C-terminal PID Domain of Fe65L1 Complexed with the Cytoplasmic Tail of APP Reveals a Novel Peptide Binding Mode | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 2, peptide from Amyloid beta A4 protein | | Authors: | Li, H, Koshiba, S, Tochio, N, Watanabe, S, Harada, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
2DVS
 
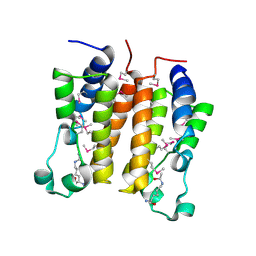 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DPC
 
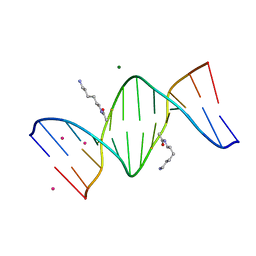 | | Crystal Structure of d(CGCGAATXCGCG) Where X is 5-(N-aminohexyl)carbamoyl-2'-O-methyluridine | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, COBALT (II) ION, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*(OMU)P*DCP*DGP*DCP*DG)-3'), ... | | Authors: | Juan, E.C.M, Kondo, J, Kurihara, T, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
2DVR
 
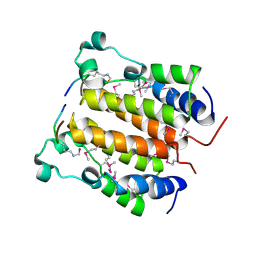 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVQ
 
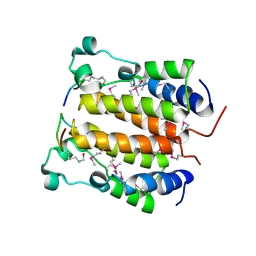 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | Bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2RRF
 
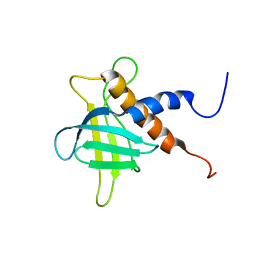 | | The solution structure of the C-terminal region of Zinc finger FYVE domain-containing protein 21 | | Descriptor: | Zinc finger FYVE domain-containing protein 21 | | Authors: | Koshiba, S, Tomizawa, T, Hayashi, F, Tochio, N, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-08-03 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ZF21 protein, a regulator of the disassembly of focal adhesions and cancer metastasis, contains a novel noncanonical pleckstrin homology domain
J.Biol.Chem., 286, 2011
|
|
2DP7
 
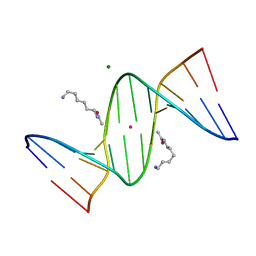 | | Crystal Structure of D(CGCGAATXCGCG) Where X is 5-(N-aminohexyl)carbamoyl-2'-deoxyuridine | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*DUP*DCP*DGP*DCP*DG)-3'), MAGNESIUM ION, ... | | Authors: | Juan, E.C.M, Kondo, J, Kurihara, T, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
2DPB
 
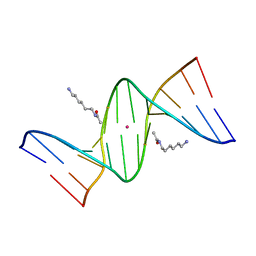 | | Crystal Structure of d(CGCGAATXCGCG) Where X is 5-(N-aminohexyl)carbamoyl-2'-deoxyuridine | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*DUP*DCP*DGP*DCP*DG)-3'), POTASSIUM ION | | Authors: | Juan, E.C.M, Kondo, J, Kurihara, T, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
1L4V
 
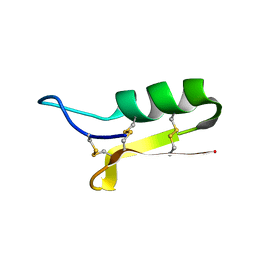 | | SOLUTION STRUCTURE OF SAPECIN | | Descriptor: | Sapecin | | Authors: | Hanzawa, H, Iwai, H, Takeuchi, K, Kuzuhara, T, Komano, H, Kohda, D, Inagaki, F, Natori, S, Arata, Y, Shimada, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-03-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | 1H nuclear magnetic resonance study of the solution conformation of an antibacterial protein, sapecin.
FEBS Lett., 269, 1990
|
|
1X0G
 
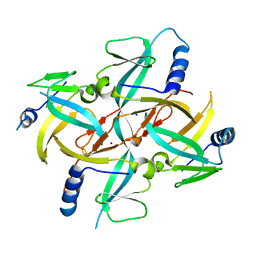 | | Crystal Structure of IscA with the [2Fe-2S] cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IscA, SODIUM ION | | Authors: | Morimoto, K, Yamashita, E, Kondou, Y, Lee, S.J, Tsukihara, T, Nakai, M. | | Deposit date: | 2005-03-22 | | Release date: | 2006-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Asymmetric IscA Homodimer with an Exposed [2Fe-2S] Cluster Suggests the Structural Basis of the Fe-S Cluster Biosynthetic Scaffold.
J.Mol.Biol., 360, 2006
|
|
4J2R
 
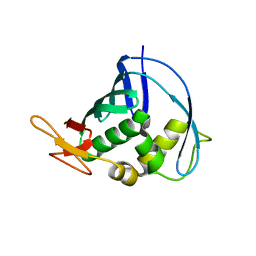 | | Middle domain of influenza A virus RNA-dependent polymerase PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Qiu, H, Tsurumura, T, Tsumori, Y, Hatakeyama, D, Kuzuhara, T, Tsuge, H. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystallization and preliminary X-ray diffraction studies of a surface mutant of the middle domain of PB2 from human influenza A (H1N1) virus
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
1WG3
 
 | | Structural analysis of yeast nucleosome-assembly factor CIA1p | | Descriptor: | Anti-silencing protein 1 | | Authors: | Padmanabhan, B, Kataoka, K, Umehara, T, Adachi, N, Yokoyama, S, Horikoshi, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Similarity between Histone Chaperone Cia1p/Asf1p and DNA-Binding Protein NF-{kappa}B
J.Biochem.(Tokyo), 138, 2005
|
|
2RV0
 
 | | Solution structures of the DNA-binding domain (ZF12) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RV6
 
 | | Solution structures of the DNA-binding domains (ZF2-ZF3-ZF4) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RV2
 
 | | Solution structures of the DNA-binding domain (ZF14) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RUT
 
 | | Solution structures of the DNA-binding domain (ZF2) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT.
J Struct Funct Genomics, 16, 2015
|
|
2RUY
 
 | | Solution structures of the DNA-binding domain (ZF10) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RUX
 
 | | Solution structures of the DNA-binding domain (ZF6) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RV4
 
 | | Solution structures of the DNA-binding domain (ZF5) of mouse immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RUW
 
 | | Solution structures of the DNA-binding domain (ZF5) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
