5WWP
 
 | | Crystal structure of Middle East respiratory syndrome coronavirus helicase (MERS-CoV nsp13) | | Descriptor: | ORF1ab, SULFATE ION, ZINC ION | | Authors: | Hao, W, Wojdyla, J.A, Zhao, R, Han, R, Das, R, Zlatev, I, Manoharan, M, Wang, M, Cui, S. | | Deposit date: | 2017-01-03 | | Release date: | 2017-07-05 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Middle East respiratory syndrome coronavirus helicase
PLoS Pathog., 13, 2017
|
|
8EJ4
 
 | | Cryo-EM structure of the active NLRP3 inflammasome disk | | Descriptor: | MAGNESIUM ION, NACHT, LRR and PYD domains-containing protein 3, ... | | Authors: | Hao, W, Le, X. | | Deposit date: | 2022-09-16 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of the active NLRP3 inflammasome disc.
Nature, 613, 2023
|
|
5X2I
 
 | |
8IMF
 
 | | Human cGAS catalytic domain bound with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xu, Y.C. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of novel cGAS inhibitors based on natural flavonoids.
Bioorg.Chem., 140, 2023
|
|
8IME
 
 | | Human cGAS catalytic domain bound with baicalin | | Descriptor: | 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xu, Y.C. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Discovery of novel cGAS inhibitors based on natural flavonoids.
Bioorg.Chem., 140, 2023
|
|
8IMG
 
 | | Human cGAS catalytic domain bound with C20 | | Descriptor: | 2-[2-hydroxy-2-oxoethyl-[3-(7-methoxy-4-methyl-2-oxidanylidene-chromen-3-yl)propanoyl]amino]ethanoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Li, J.M, Xu, Y.C. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of novel cGAS inhibitors based on natural flavonoids.
Bioorg.Chem., 140, 2023
|
|
6LRC
 
 | | Human cGAS catalytic domain bound with the inhibitor PF-06928215 | | Descriptor: | (1R,2S)-2-[(7-hydroxy-5-phenylpyrazolo[1,5-a]pyrimidine-3-carbonyl)amino]cyclohexane-1-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xu, Y.C. | | Deposit date: | 2020-01-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LRL
 
 | | Human cGAS catalytic domain bound with compound s2 | | Descriptor: | 3-[[5-(1,2,4-triazol-4-yl)-4H-1,2,4-triazol-3-yl]carbonylamino]benzoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LRJ
 
 | | Human cGAS catalytic domain bound with compound 23 | | Descriptor: | 4-[2-(2-methyl-[1,2,4]triazolo[1,5-c]quinazolin-5-yl)hydrazinyl]-4-oxidanylidene-butanoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
5ME3
 
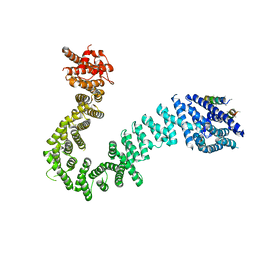 | | Structure of the Scc2 C-terminus | | Descriptor: | Scc2 unassigned sequence, Sister chromatid cohesion protein 2, unassigned sequence of Scc2 | | Authors: | Chao, W.C.H, Singleton, M.R. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the cohesin loader Scc2.
Nat Commun, 8, 2017
|
|
5N22
 
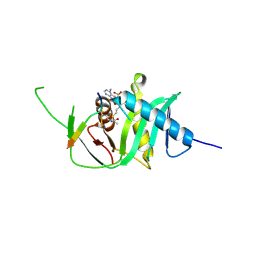 | | Structure of xEco2 acetyltransferase domain bound to K106-CoA conjugate | | Descriptor: | (2~{S})-2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]propanoic acid, GLY-ALA-LYS-LYX-ASP-GLN-TYR-PHE-LEU, XEco2 | | Authors: | Chao, W.C.H, Wade, B.O, Singleton, M.R. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of Eco1-Mediated Cohesin Acetylation.
Sci Rep, 7, 2017
|
|
5N1W
 
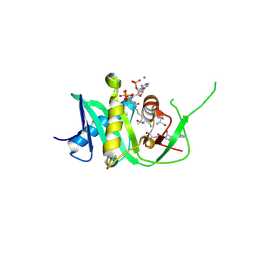 | | Structure of xEco2 acetyltransferase domain bound to K105-CoA conjugate | | Descriptor: | (2~{S})-2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]propanoic acid, ILE-GLY-ALA-LYX-LYS-ALA,ILE-GLY-ALA-LYX-LYS-ALA,ILE-GLY-ALA-LYX-LYS-ALA,ILE-GLY-ALA-LYX-LYS-ALA, MAGNESIUM ION, ... | | Authors: | Chao, W.C.H, Wade, B.O, Singleton, M.R. | | Deposit date: | 2017-02-06 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Eco1-Mediated Cohesin Acetylation.
Sci Rep, 7, 2017
|
|
5N1U
 
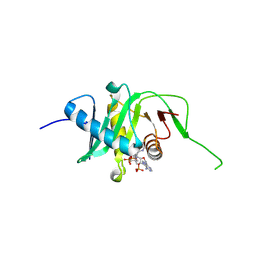 | |
6LRI
 
 | | Human cGAS catalytic domain bound with compound 17 | | Descriptor: | 3-[5-(2-hydroxy-2-oxoethyl)-3-oxidanylidene-[1,2,4]triazino[2,3-a]benzimidazol-2-yl]propanoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LRE
 
 | | Human cGAS catalytic domain bound with compound 3 | | Descriptor: | 1,3-bis(oxidanylidene)benzo[de]isoquinoline-6,7-dicarboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LRK
 
 | | Human cGAS catalytic domain bound with compound 40 | | Descriptor: | (3R)-1-pyrrolo[1,2-a]quinoxalin-4-ylpiperidine-3-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
8BD1
 
 | |
1HHV
 
 | | SOLUTION STRUCTURE OF VIRUS CHEMOKINE VMIP-II | | Descriptor: | VIRUS CHEMOKINE VMIP-II | | Authors: | Shao, W, Fernandez, E, Navenot, J.M, Wilken, J, Thompson, D.A, Pepiper, S, Schweitzer, B.I, Lolis, E. | | Deposit date: | 1998-12-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | CCR2 and CCR5 receptor-binding properties of herpesvirus-8
vMIP-II based on sequence analysis and its solution structure
Eur.J.Biochem., 268, 2001
|
|
8GZE
 
 | | Crystal Structure of human METTL9-SAH-SLC39A7 peptide complex | | Descriptor: | Protein-L-histidine N-pros-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, Zinc transporter SLC39A7 | | Authors: | Zhao, W.T, Li, H.T. | | Deposit date: | 2022-09-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular basis for protein histidine N1-specific methylation of the "His-x-His" motifs by METTL9.
Cell Insight, 2, 2023
|
|
8GZF
 
 | | Crystal Structure of METTL9-SAH | | Descriptor: | Protein-L-histidine N-pros-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zhao, W.T, Li, H.T. | | Deposit date: | 2022-09-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for protein histidine N1-specific methylation of the "His-x-His" motifs by METTL9.
Cell Insight, 2, 2023
|
|
1MI2
 
 | | SOLUTION STRUCTURE OF MURINE MACROPHAGE INFLAMMATORY PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN-2 | | Authors: | Shao, W, Jerva, L.F, West, J, Lolis, E, Schweitzer, B.I. | | Deposit date: | 1997-10-24 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of murine macrophage inflammatory protein-2.
Biochemistry, 37, 1998
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XK8
 
 | | Cryo-EM structure of the Neuromedin U receptor 2 (NMUR2) in complex with G Protein and its endogeneous Peptide-Agonist NMU25 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, W, Wenru, Z, Mu, W, Minmin, L, Shutian, C, Tingting, T, Gisela, S, Holger, W, Albert, B, Cuiying, Y, Xiaojing, C, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and activation of neuromedin U receptor 2.
Nat Commun, 13, 2022
|
|
4M70
 
 | |
