8K6N
 
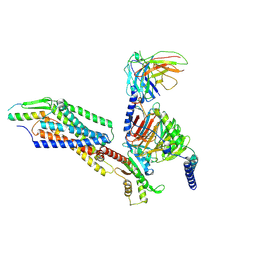 | | Structural complex of neuropeptide Y receptor 2 | | 分子名称: | C-flanking peptide of NPY, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Shen, S.Y, Zhao, C, Shao, Z.H. | | 登録日 | 2023-07-25 | | 公開日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural complex of neuropeptide Y receptor 2
To Be Published
|
|
8K6O
 
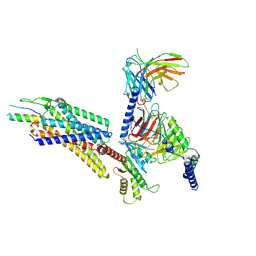 | | Structural complex of neuropeptide Y receptor 1 | | 分子名称: | C-flanking peptide of NPY, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Shen, S.Y, Zhao, C, Shao, Z.H. | | 登録日 | 2023-07-25 | | 公開日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural complex of neuropeptide Y receptor 1
To Be Published
|
|
7D90
 
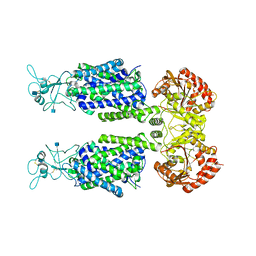 | | human potassium-chloride co-transporter KCC3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | 著者 | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | 登録日 | 2020-10-12 | | 公開日 | 2020-12-30 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7FEE
 
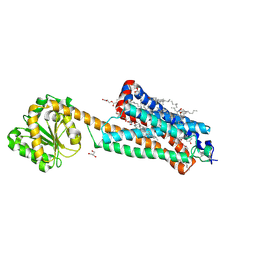 | | Crystal structure of the allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, ... | | 著者 | Wang, X, Zhao, C, Shao, Z. | | 登録日 | 2021-07-19 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
5N3U
 
 | | The structure of the complex of CpcE and CpcF of phycocyanin lyase from Nostoc sp. PCC7120 | | 分子名称: | Phycocyanobilin lyase subunit alpha, Phycocyanobilin lyase subunit beta | | 著者 | Hoeppner, A, Zhao, C, Xu, Q.-Z, Gaertner, W, Scheer, H, Zhao, K.-H. | | 登録日 | 2017-02-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structures and enzymatic mechanisms of phycobiliprotein lyases CpcE/F and PecE/F.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8K6M
 
 | | Structural complex of neuropeptide Y receptor 1 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | 著者 | Shen, S.Y, Zhao, C, Shao, Z.H. | | 登録日 | 2023-07-25 | | 公開日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural complex of neuropeptide Y receptor 1
To Be Published
|
|
7D99
 
 | | human potassium-chloride co-transporter KCC4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | 著者 | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | 登録日 | 2020-10-12 | | 公開日 | 2020-12-30 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D8Z
 
 | | human potassium-chloride co-transporter KCC2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | 著者 | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | 登録日 | 2020-10-11 | | 公開日 | 2020-12-30 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7BOR
 
 | | Structure of Pseudomonas aeruginosa CoA-bound OdaA | | 分子名称: | COENZYME A, Probable enoyl-CoA hydratase/isomerase | | 著者 | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | 登録日 | 2020-03-19 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
7Y8W
 
 | | Crystal structure of DLC-1/SAO-1 complex | | 分子名称: | Dynein light chain 1, cytoplasmic, GLYCEROL, ... | | 著者 | Yan, H, Zhao, C, Wei, Z, Yu, C. | | 登録日 | 2022-06-24 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Interaction between DLC-1 and SAO-1 facilitates CED-4 translocation during apoptosis in the Caenorhabditis elegans germline.
Cell Death Discov, 8, 2022
|
|
5YKJ
 
 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | 分子名称: | GLYCEROL, Peroxiredoxin PRX1, mitochondrial, ... | | 著者 | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | 登録日 | 2017-10-14 | | 公開日 | 2018-10-24 | | 最終更新日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
To be published
|
|
6JAU
 
 | | The complex structure of Pseudomonas aeruginosa MucA/MucB. | | 分子名称: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | 著者 | Li, T, He, L.H, Li, C.C, Liu, L, Peng, C.T, Shen, Y.L, Qin, X.F, Xiao, Q.J, Zhu, Y.B, Song, Y.J, Zhao, N.l, Zhao, C, Yang, J, Mu, X.Y, Huang, Q, Bao, R. | | 登録日 | 2019-01-25 | | 公開日 | 2020-01-29 | | 最終更新日 | 2020-08-19 | | 実験手法 | X-RAY DIFFRACTION (1.905 Å) | | 主引用文献 | Molecular basis of the lipid-induced MucA-MucB dissociation in Pseudomonas aeruginosa.
Commun Biol, 3, 2020
|
|
5YKW
 
 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | 分子名称: | Thioredoxin-3, mitochondrial, peptide THR-PRO-VAL-CYS-THR-THR-GLU-VAL | | 著者 | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | 登録日 | 2017-10-16 | | 公開日 | 2018-10-24 | | 最終更新日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
to be published
|
|
1YT9
 
 | | HIV Protease with oximinoarylsulfonamide bound | | 分子名称: | (S)-N-((2S,3R)-3-HYDROXY-4-(4-((E)-(HYDROXYIMINO)METHYL)-N-ISOBUTYLPHENYLSULFONAMIDO)-1-PHENYLBUTAN-2-YL)-3-METHYL-2-(3 -((2-METHYLTHIAZOL-4-YL)METHYL)-2-OXOIMIDAZOLIDIN-1-YL)BUTANAMIDE, Pol polyprotein | | 著者 | Yeung, C.M, Klein, L.L, Flentge, C.A, Randolph, J.T, Zhao, C, Sun, M, Dekhtyar, T, Stoll, V.S, Kempf, D.J. | | 登録日 | 2005-02-10 | | 公開日 | 2005-04-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Oximinoarylsulfonamides as potent HIV protease inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8GHF
 
 | | cryo-EM structure of hSlo1 in plasma membrane vesicles | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL, ... | | 著者 | Tao, X, Zhao, C, MacKinnon, R. | | 登録日 | 2023-03-10 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Membrane protein isolation and structure determination in cell-derived membrane vesicles.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GHG
 
 | |
8GH9
 
 | |
6IGB
 
 | | the structure of Pseudomonas aeruginosa Periplasmic gluconolactonase, PpgL | | 分子名称: | ACETATE ION, Periplasmic gluconolactonase, PpgL, ... | | 著者 | Song, Y.J, Shen, Y.L, Wang, K.L, Li, T, Zhu, Y.B, Li, C.C, He, L.H, Zhao, N.L, Zhao, C, Yang, J, Huang, Q, Mu, X.Y. | | 登録日 | 2018-09-25 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Structural and Functional Insights into PpgL, a Metal-Independent beta-Propeller Gluconolactonase That Contributes toPseudomonas aeruginosaVirulence.
Infect.Immun., 87, 2019
|
|
5QD2
 
 | | Crystal structure of BACE complex with BMC017 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(methoxymethyl)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCS
 
 | | Crystal structure of BACE complex with BMC024 | | 分子名称: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDC
 
 | | Crystal structure of BACE complex with BMC019 hydrolyzed | | 分子名称: | (4S)-4-[(1R)-1,2-dihydroxyethyl]-N,N-dimethyl-2-oxo-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaene-19-carboxamide, Beta-secretase 1, GLYCEROL | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCW
 
 | | Crystal structure of BACE complex with BMC021 | | 分子名称: | (2R,4S)-N-butyl-4-[(2S,5S,7R)-2,7-dimethyl-3,15-dioxo-1,4-diazacyclopentadecan-5-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDA
 
 | | Crystal structure of BACE complex with BMC013 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-18-methoxy-3,15,17-triazatricyclo[14.3.1.1~6,10~]henicosa-1(20),6(21),7,9,16,18-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD7
 
 | | Crystal structure of BACE complex with BMC014 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({1-[3-(1-methylethyl)phenyl]cyclopropyl}amino)ethyl]-19-(methoxymethyl)-11-oxa-3,16-diazatric yclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCQ
 
 | | Crystal structure of BACE complex with BMC025 | | 分子名称: | (2R,4S,5S)-N-butyl-4-hydroxy-2,7-dimethyl-5-{[N-(4-methylpentanoyl)-L-methionyl]amino}octanamide, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
