6IEV
 
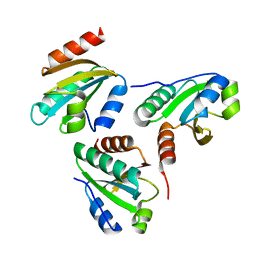 | | Crystal structure of a designed protein | | Descriptor: | Designed protein | | Authors: | Han, M, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Selection and analyses of variants of a designed protein suggest importance of hydrophobicity of partially buried sidechains for protein stability at high temperatures.
Protein Sci., 28, 2019
|
|
1GGZ
 
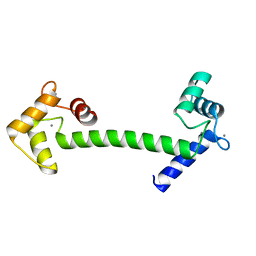 | | CRYSTAL STRUCTURE OF THE CALMODULIN-LIKE PROTEIN (HCLP) FROM HUMAN EPITHELIAL CELLS | | Descriptor: | CALCIUM ION, CALMODULIN-RELATED PROTEIN NB-1 | | Authors: | Han, B.-G, Han, M, Sui, H, Yaswen, P, Walian, P.J, Jap, B.K. | | Deposit date: | 2000-10-13 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human calmodulin-like protein: insights into its functional role.
FEBS Lett., 521, 2002
|
|
5GS5
 
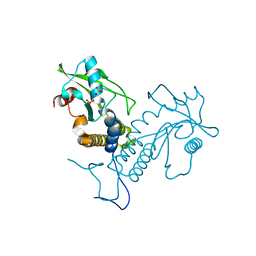 | | Crystal structure of apo rat STING | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of apo ratSTING
To Be Published
|
|
5GRM
 
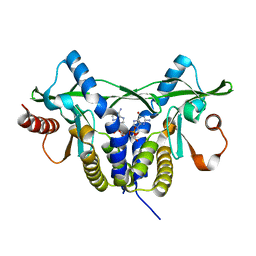 | | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP) | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-11 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP)
To Be Published
|
|
1HUX
 
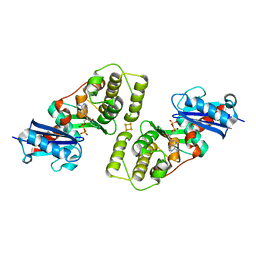 | | CRYSTAL STRUCTURE OF THE ACIDAMINOCOCCUS FERMENTANS (R)-2-HYDROXYGLUTARYL-COA DEHYDRATASE COMPONENT A | | Descriptor: | ACTIVATOR OF (R)-2-HYDROXYGLUTARYL-COA DEHYDRATASE, ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER | | Authors: | Locher, K.P, Hans, M, Yeh, A.P, Schmid, B, Buckel, W, Rees, D.C. | | Deposit date: | 2001-01-04 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Acidaminococcus fermentans 2-hydroxyglutaryl-CoA dehydratase component A.
J.Mol.Biol., 307, 2001
|
|
1G4R
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN 1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
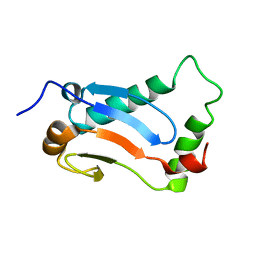
1G4M
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
2MUK
 
 | |
3UKR
 
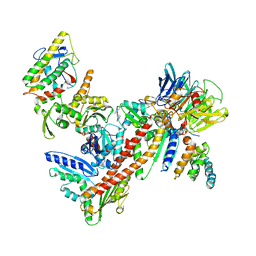 | | Crystal structure of Bos taurus Arp2/3 complex with bound inhibitor CK-666 | | Descriptor: | 2-fluoro-N-[2-(2-methyl-1H-indol-3-yl)ethyl]benzamide, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | Nolen, B.J, Han, M. | | Deposit date: | 2011-11-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural characterization and computer-aided optimization of a small-molecule inhibitor of the Arp2/3 complex, a key regulator of the actin cytoskeleton.
Chemmedchem, 7, 2012
|
|
8OWJ
 
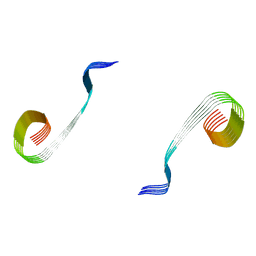 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OVK
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L1 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWD
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWK
 
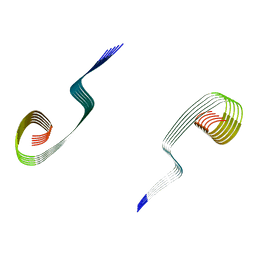 | | Lipidic amyloid-beta(1-40) fibril - polymorph L3-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OVM
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWE
 
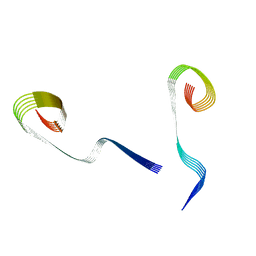 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
3UKU
 
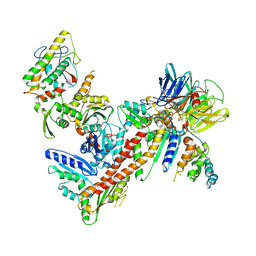 | | Structure of Arp2/3 complex with bound inhibitor CK-869 | | Descriptor: | (2S)-2-(3-bromophenyl)-3-(2,4-dimethoxyphenyl)-1,3-thiazolidin-4-one, ACTIN-LIKE PROTEIN 2, ACTIN-LIKE PROTEIN 3, ... | | Authors: | Nolen, B.J, Han, M. | | Deposit date: | 2011-11-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Arp2/3 complex with bound inhibitor CK-869
To be Published
|
|
3ULE
 
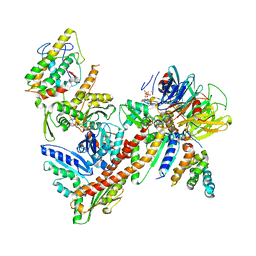 | |
