3CYG
 
 | |
3UP8
 
 | | Crystal structure of a putative 2,5-diketo-D-gluconic acid reductase B | | Descriptor: | ACETATE ION, Putative 2,5-diketo-D-gluconic acid reductase B | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a putative 2,5-diketo-D-gluconic acid reductase B
To be Published
|
|
3CXJ
 
 | |
3V4C
 
 | | Crystal structure of a semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Aldehyde dehydrogenase (NADP+) | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of a semialdehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3R79
 
 | |
1DQ8
 
 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG AND COA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-HYDROXY-3-METHYL-GLUTARIC ACID, COENZYME A, ... | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
3TFW
 
 | |
2IMR
 
 | |
1S0B
 
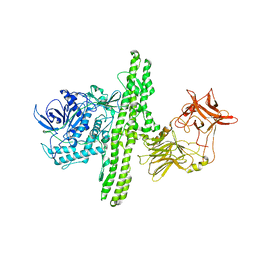 | | Crystal structure of botulinum neurotoxin type B at pH 4.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1S0D
 
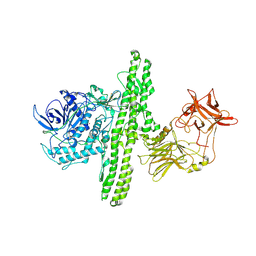 | | Crystal structure of botulinum neurotoxin type B at pH 5.5 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
2IMG
 
 | | Crystal structure of dual specificity protein phosphatase 23 from Homo sapiens in complex with ligand malate ion | | Descriptor: | D-MALATE, Dual specificity protein phosphatase 23 | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of human dual specificity protein phosphatase 23, VHZ, enzyme-substrate/product complex.
J.Biol.Chem., 283, 2008
|
|
1S0G
 
 | |
1D1U
 
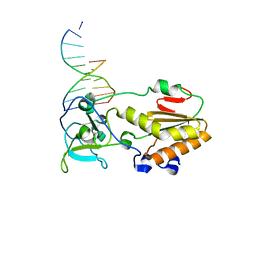 | | USE OF AN N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE TO FACILITATE CRYSTALLIZATION AND ANALYSIS OF A PSEUDO-16-MER DNA MOLECULE CONTAINING G-A MISPAIRS | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*CP*AP*CP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*GP*TP*G)-3'), PROTEIN (REVERSE TRANSCRIPTASE) | | Authors: | Cote, M.L, Yohannan, S, Georgiadis, M.M. | | Deposit date: | 1999-09-21 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of an N-terminal fragment from moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing G-A mispairs.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2ICS
 
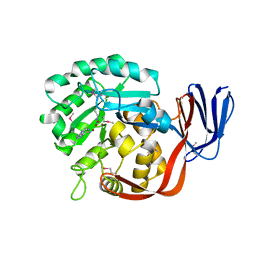 | | Crystal structure of an adenine deaminase | | Descriptor: | ADENINE, Adenine Deaminase, ZINC ION | | Authors: | Sugadev, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an adenine deaminase
TO BE PUBLISHED
|
|
2K0U
 
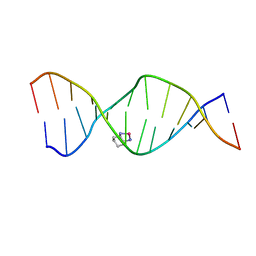 | | High Resolution Solution NMR Structures of Oxaliplatin-DNA Adduct | | Descriptor: | CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), DNA (5'-D(*DCP*DCP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DTP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DGP*DG)-3') | | Authors: | Bhattacharyya, D, King, C.L, Chaney, S.G, Campbell, S.L. | | Deposit date: | 2008-02-15 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Flanking Bases Influence the Nature of DNA Distortion by Platinum 1,2-Intrastrand (GG) Cross-Links.
Plos One, 6, 2011
|
|
2K0V
 
 | | High Resolution Solution NMR Structures of Undamaged DNA Dodecamer Duplex | | Descriptor: | DNA (5'-D(*DCP*DCP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DTP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DGP*DG)-3') | | Authors: | Bhattacharyya, D, King, C.L, Chaney, S.G, Campbell, S.L. | | Deposit date: | 2008-02-15 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Flanking Bases Influence the Nature of DNA Distortion by Platinum 1,2-Intrastrand (GG) Cross-Links.
Plos One, 6, 2011
|
|
2JUV
 
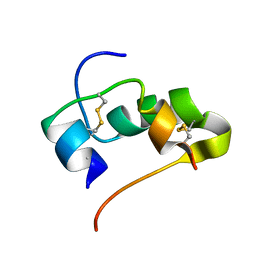 | | AbaA3-DKP-insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Huang, K, Chan, S, Hua, Q, Chu, Y, Wang, R, Klaproth, B, Jia, W, Whittaker, J, De Meyts, P, Nakagawa, S.H, Steiner, D.F, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2007-09-05 | | Release date: | 2007-10-16 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The A-chain of Insulin Contacts the Insert Domain of the Insulin Receptor: PHOTO-CROSS-LINKING AND MUTAGENESIS OF A DIABETES-RELATED CREVICE.
J.Biol.Chem., 282, 2007
|
|
3US8
 
 | | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Isocitrate dehydrogenase [NADP], SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
2ABQ
 
 | |
3CLK
 
 | |
3CO4
 
 | |
1U05
 
 | |
1TXZ
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-06 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme.
Protein Sci., 14, 2005
|
|
2AHW
 
 | | Crystal Structure of Acyl-CoA transferase from E. coli O157:H7 (YdiF)-thioester complex with CoA- 2 | | Descriptor: | COENZYME A, putative enzyme YdiF | | Authors: | Rangarajan, E.S, Li, Y, Ajamian, E, Iannuzzi, P, Kernaghan, S.D, Fraser, M.E, Cylger, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
J.Biol.Chem., 280, 2005
|
|
3CVG
 
 | |
