3K6U
 
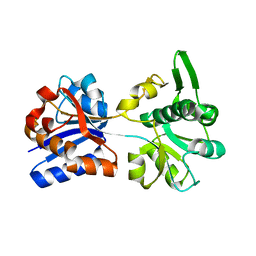 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1Z15
 
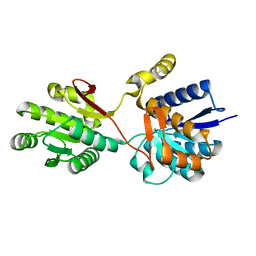 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein in superopen form | | Descriptor: | Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
1ZUG
 
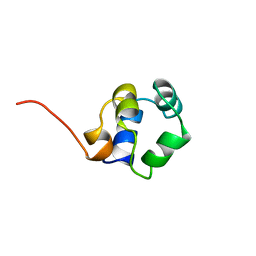 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
1Z17
 
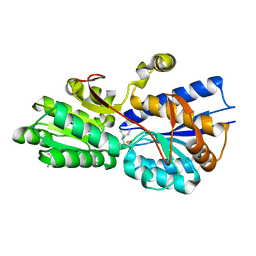 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound ligand isoleucine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOLEUCINE, Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
3SLN
 
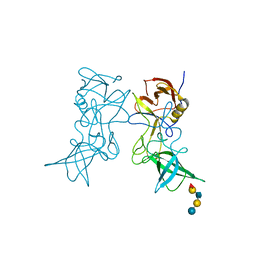 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to H pentasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
1Z8F
 
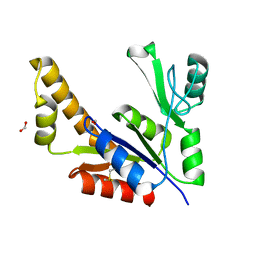 | | Guanylate Kinase Double Mutant A58C, T157C from Mycobacterium tuberculosis (Rv1389) | | Descriptor: | FORMIC ACID, Guanylate kinase | | Authors: | Chan, S, Sawaya, M.R, Choi, B, Zocchi, G, Perry, L.J, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-03-30 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Guanylate Kinase from Mycobacterium tuberculosis (Rv1389)
To be Published
|
|
3THF
 
 | |
3SKX
 
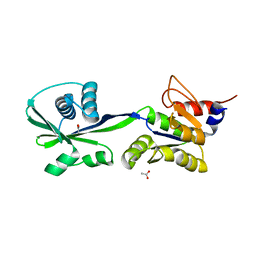 | | Crystal structure of the ATP binding domain of Archaeoglobus fulgidus COPB | | Descriptor: | ACETATE ION, Copper-exporting P-type ATPase B | | Authors: | Jayakanthan, S, Roberts, S.A, Weichsel, A, Arguello, J.M, McEvoy, M.M. | | Deposit date: | 2011-06-23 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Conformations of the apo-, substrate-bound and phosphate-bound ATP-binding domain of the Cu(II) ATPase CopB illustrate coupling of domain movement to the catalytic cycle.
Biosci.Rep., 32, 2012
|
|
3SPJ
 
 | |
2MDA
 
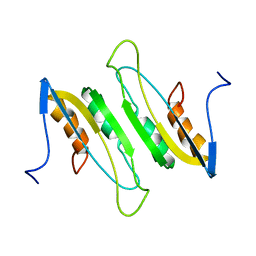 | |
3SEJ
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to Secretor Lewis HBGA (LeB) | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SLD
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to A trisaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SKB
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) | | Descriptor: | Capsid | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SJP
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) | | Descriptor: | Capsid, ZINC ION | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
1EYG
 
 | |
3Q4H
 
 | | Crystal structure of the Mycobacterium smegmatis EsxGH complex (MSMEG_0620-MSMEG_0621) | | Descriptor: | Low molecular weight protein antigen 7, Pe family protein | | Authors: | Chan, S, Harris, L, Kuo, E, Ahn, C, Zhou, T.T, Nguyen, L, Shin, A, Sawaya, M.R, Cascio, D, Arbing, M.A, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-12-23 | | Release date: | 2011-01-26 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
4JFA
 
 | | Crystal Structure of Plasmodium falciparum Tryptophanyl-tRNA synthetase | | Descriptor: | BETA-MERCAPTOETHANOL, POTASSIUM ION, TRYPTOPHAN, ... | | Authors: | Khan, S, Garg, A, Manickam, Y, Sharma, A. | | Deposit date: | 2013-02-28 | | Release date: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An appended domain results in an unusual architecture for malaria parasite tryptophanyl-tRNA synthetase
Plos One, 8, 2013
|
|
2K43
 
 | |
4PDQ
 
 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neomycin analog | | Descriptor: | (2S)-4-amino-N-{(1R,2S,3R,4R,5S)-5-amino-3-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy }-4-[(2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-galactopyranosyl)oxy]-2-hydroxycyclohexyl}-2-hydroxybutanamide, MAGNESIUM ION, RNA (5'-*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C-3') | | Authors: | Hanessian, S, Saavedra, O.M, Vilchis-Reyes, M.A, Maianti, J.P, Kanazawa, H, Dozzo, P, Feeney, L.A, Armstrong, E.S, Kondo, J. | | Deposit date: | 2014-04-21 | | Release date: | 2015-01-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, broad spectrum antibacterial activity, and X-ray co-crystal structure of the decoding bacterial ribosomal A-site with 4'-deoxy-4'-fluoro neomycin analogs
Chem Sci, 5, 2014
|
|
2O8O
 
 | |
1KAW
 
 | | STRUCTURE OF SINGLE STRANDED DNA BINDING PROTEIN (SSB) | | Descriptor: | SINGLE-STRANDED DNA BINDING PROTEIN | | Authors: | Raghunathan, S, Waksman, G. | | Deposit date: | 1996-12-06 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2K69
 
 | |
2K68
 
 | |
4L2W
 
 | |
6J42
 
 | | Crystal Structure of Wild Type KatB, a manganese catalase from Anabaena | | Descriptor: | Alr3090 protein, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Bihani, S.C, Chakravarty, D, Ballal, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Novel molecular insights into the anti-oxidative stress response and structure-function of a salt-inducible cyanobacterial Mn-catalase.
Plant Cell Environ, 42, 2019
|
|
