3UDF
 
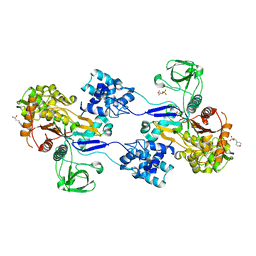 | | Crystal structure of Apo PBP1a from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UDI
 
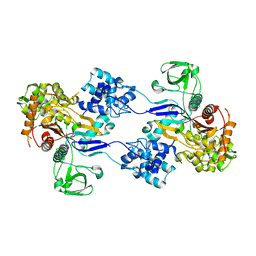 | |
3UDX
 
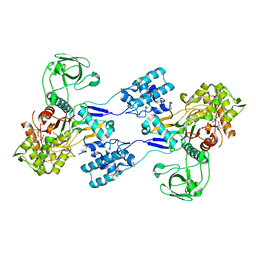 | | Crystal structure of Acinetobacter baumannii PBP1a in complex with Imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UE0
 
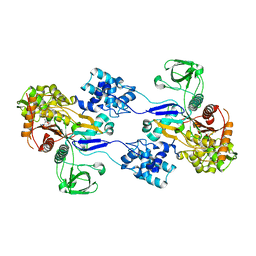 | | Crystal structure of Acinetobacter baumannii PBP1a in complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
4OON
 
 | | Crystal structure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) | | Descriptor: | (4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid, Penicillin-binding protein 1A | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
4OOL
 
 | | Crystal structure of PBP3 in complex with compound 14 ((2E)-2-({[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}acetyl]amino}-3-oxopropyl]oxy}imino)pentanedioic acid) | | Descriptor: | (2E)-2-({[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}acetyl]amino}-3-oxopropyl]oxy}imino)pentanedioic acid, Cell division protein FtsI [Peptidoglycan synthetase] | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
8WWV
 
 | | 1-naphthylamine GS in complex with ADP and MetSox-P | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Zhang, S.T, Zhou, N.Y. | | Deposit date: | 2023-10-26 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the 1-naphthylamine biodegradation pathway reveals a glutamine synthetase-like protein that catalyzes 1-naphthylamine glutamylation
To Be Published
|
|
8WWU
 
 | | 1-naphthylamine GS in complex with AMP PNP | | Descriptor: | Glutamine synthetase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Zhang, S.T, Zhou, N.Y. | | Deposit date: | 2023-10-26 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the 1-naphthylamine biodegradation pathway reveals a glutamine synthetase-like protein that catalyzes 1-naphthylamine glutamylation
To Be Published
|
|
3U2G
 
 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1HNB
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1HNC
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
8WR4
 
 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex) | | Descriptor: | CbCas9 effector-1, DNA (62-MER), MAGNESIUM ION, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
5WLL
 
 | |
8WMM
 
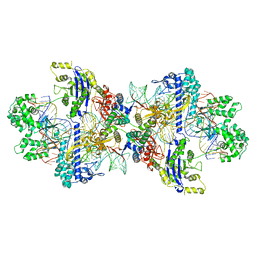 | | Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary) | | Descriptor: | MAGNESIUM ION, NTS, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMH
 
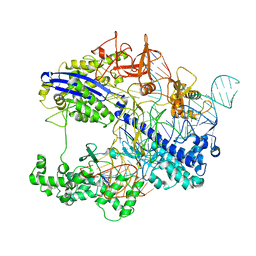 | | Structure of CbCas9 bound to 6-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMN
 
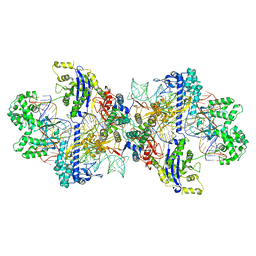 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary) | | Descriptor: | DNA (62-MER), MAGNESIUM ION, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMV
 
 | | The structure of PSI-14CAC complex at stationary growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8WMJ
 
 | | structure of PSI-11CAC complex at Logrithmic growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8WM6
 
 | | The structure of PSI-CAC(L-14)of R.salina at 2.7 angstroms resolution | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8WMW
 
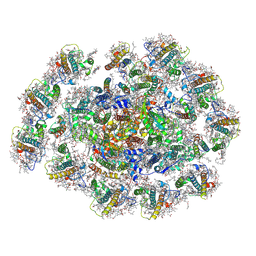 | | The structure of PSI-11CAC at the stationary growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8WNW
 
 | | the structure of PsaQ | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, PsaQ | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
1HZ3
 
 | | ALZHEIMER'S DISEASE AMYLOID-BETA PEPTIDE (RESIDUES 10-35) | | Descriptor: | A-BETA AMYLOID | | Authors: | Zhang, S, Iwata, K, Lachenmann, M.J, Peng, J.W, Li, S, Stimson, E.R, Lu, Y, Felix, A.M, Maggio, J.E, Lee, J.P. | | Deposit date: | 2001-01-23 | | Release date: | 2001-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Alzheimer's peptide a beta adopts a collapsed coil structure in water.
J.Struct.Biol., 130, 2000
|
|
3UEJ
 
 | |
3UGL
 
 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | PHOSPHATE ION, Proteine kinase C delta type, ZINC ION, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.357 Å) | | Cite: | Structural and Functional Characterization of an Anesthetic Binding Site in the Second Cysteine-Rich Domain of Protein Kinase Cdelta
Biophys.J., 103, 2012
|
|
3UFF
 
 | |
