6FAL
 
 | |
4XPW
 
 | | Crystal structures of Leu114F mutant | | Descriptor: | Bacteriohemerythrin, FE (II) ION, GLYCEROL | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
6EZ0
 
 | | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding | | Descriptor: | RNA (27-MER) | | Authors: | Erat, M.C, Besic, E, Oberhuber, M, Johannsen, S, Sigel, R.K.O. | | Deposit date: | 2017-11-13 | | Release date: | 2018-01-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding.
J. Biol. Inorg. Chem., 23, 2018
|
|
6F7F
 
 | |
6F7G
 
 | |
6F9K
 
 | |
4XPY
 
 | | Crystal structure of hemerythrin : L114Y mutant | | Descriptor: | Bacteriohemerythrin, FE (II) ION, GLYCEROL | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
3K2G
 
 | | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, MAGNESIUM ION, Resiniferatoxin-binding, ... | | Authors: | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides
To be Published
|
|
3E61
 
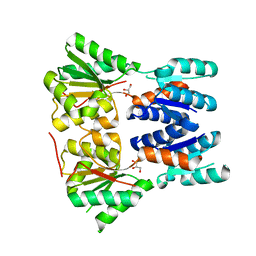 | | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | GLYCEROL, Putative transcriptional repressor of ribose operon | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-14 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus
To be Published
|
|
7NQA
 
 | | Crystal structure of Nucleoporin-98 nanobody MS98-6 complex solved at 2.2A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-Nup98 Nanobody MS98-6, Nuclear pore complex protein Nup98-Nup96, ... | | Authors: | Sola-Colom, M, Trakhanov, S, Goerlich, D. | | Deposit date: | 2021-03-01 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A checkpoint function for Nup98 in nuclear pore formation suggested by novel inhibitory nanobodies.
Embo J., 2024
|
|
4Y9H
 
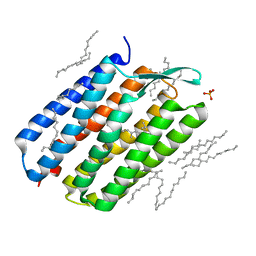 | | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, DECANE, DODECANE, ... | | Authors: | Saiki, H, Sugiyama, S, Kakinouchi, K, Kawatake, S, Hanashima, S, Matsumori, N, Murata, M. | | Deposit date: | 2015-02-17 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles
To Be Published
|
|
3EEQ
 
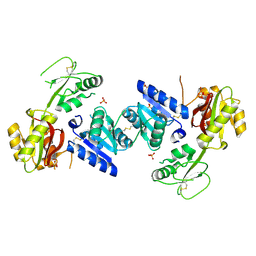 | | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus | | Descriptor: | SULFATE ION, putative Cobalamin biosynthesis protein G homolog | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-05 | | Release date: | 2008-09-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus
To be Published
|
|
3C8T
 
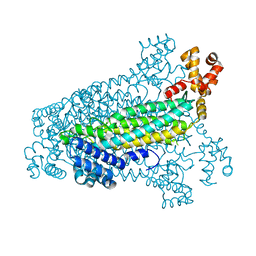 | | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1 | | Descriptor: | Fumarate lyase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-13 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1.
To be Published
|
|
4X1M
 
 | |
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | Authors: | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
5DJA
 
 | | Fc Heterodimer Design 9.1 Y407M + T366I | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
3DXI
 
 | |
3DZ1
 
 | |
5DA3
 
 | | Crystal structure of PTK6 Kinase domain with inhibitor | | Descriptor: | (2-chloro-4-{[6-cyclopropyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}phenyl)(morpholin-4-yl)methanone, GLYCEROL, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Battula, S.K, Vadivelu, S, Tyagi, R, Gosu, R. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structures of PTK6: With Dasatinib at 2.24 angstrom , with novel imidazo[1,2-a]pyrazin-8-amine derivative inhibitor at 1.70 angstrom resolution
Biochem. Biophys. Res. Commun., 482, 2017
|
|
8OV5
 
 | |
3B89
 
 | |
5IXM
 
 | | The LPS Transporter LptDE from Yersinia pestis, core complex | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Botos, I, Mayclin, S.J, McCarthy, J.G, Buchanan, S.K. | | Deposit date: | 2016-03-23 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens.
Structure, 24, 2016
|
|
3MKC
 
 | |
3M3M
 
 | | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5] | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Bagaria, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5]
To be Published
|
|
3KOL
 
 | |
