3H1K
 
 | | Chicken cytochrome BC1 complex with ZN++ and an iodinated derivative of kresoxim-methyl bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Berry, E.A, Zhang, Z, Bellamy, H.D, Huang, L.S. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Crystallographic location of two Zn(2+)-binding sites in the avian cytochrome bc(1) complex
Biochim.Biophys.Acta, 1459, 2000
|
|
6B7Y
 
 | | Cryo-EM structure of human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF6
 
 | | Cryo-EM structure of human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B3Q
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-22 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
1X11
 
 | | X11 PTB DOMAIN | | Descriptor: | 13-MER PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
6BFC
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-27 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Z
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain | | Descriptor: | FAB H11 heavy chain, FAB H11 light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-05 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF9
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
7K0N
 
 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6M4Q
 
 | | Cytochrome P450 monooxygenase StvP2 substrate-free structure | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
5BX1
 
 | | Crystal Structure of PRL-1 complex with compound analogy 3 | | Descriptor: | 3-(5,6-dimethyl-2H-isoindol-2-yl)-N'-[(E)-furan-2-ylmethylidene]benzohydrazide, Protein tyrosine phosphatase type IVA 1, SULFATE ION | | Authors: | Liu, S, Bai, Y, Zhang, Z. | | Deposit date: | 2015-06-08 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of PRL-1 complex with compound analogy 3
To Be Published
|
|
5GPI
 
 | |
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
7K56
 
 | | Structure of VCP dodecamer purified from H1299 cells | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
7K59
 
 | | Structure of apo VCP hexamer generated from bacterially recombinant VCP/p97 | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
7YWX
 
 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7K57
 
 | | Structure of apo VCP dodecamer generated from bacterially recombinant VCP/p97 | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
8HKF
 
 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, Z.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
5ZBA
 
 | | Crystal structure of Rtt109-Asf1-H3-H4-CoA complex | | Descriptor: | COENZYME A, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
1JOT
 
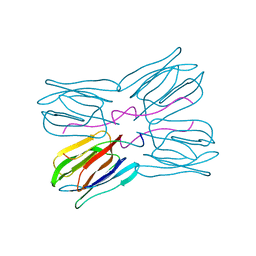 | | STRUCTURE OF THE LECTIN MPA COMPLEXED WITH T-ANTIGEN DISACCHARIDE | | Descriptor: | AGGLUTININ, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Lee, X, Thompson, A, Zhang, Z, Hoa, T.-T, Biesterfeldt, J, Ogata, C, Xu, L, Johnston, R.A.Z, Young, N.M. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of Maclura pomifera agglutinin and the T-antigen disaccharide, Galbeta1,3GalNAc.
J.Biol.Chem., 273, 1998
|
|
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
8IDN
 
 | | Cryo-EM structure of RBD/E77-Fab complex | | Descriptor: | E77 Fab heavy chain, E77 Fab light chain, Spike protein S1, ... | | Authors: | Lu, D.F, Zhang, Z.C. | | Deposit date: | 2023-02-13 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structure of the RBD-E77 Fab complex reveals neutralization and immune escape of SARS-CoV-2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4DGX
 
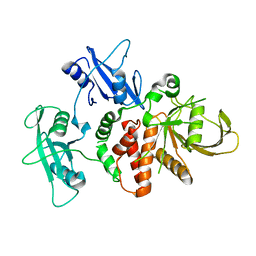 | | LEOPARD Syndrome-Associated SHP2/Y279C mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
4L0C
 
 | | Crystal structure of the N-Fopmylmaleamic acid deformylase Nfo(S94A) from Pseudomonas putida S16 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Deformylase | | Authors: | Chen, D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | Deposit date: | 2013-05-31 | | Release date: | 2014-06-04 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the N-Fopmylmaleamic acid deformylase Nfo from Pseudomonas putida S16
To be Published
|
|
