8GJL
 
 | | multi-drug efflux pump RE-CmeB bound with Ciprofloxacin | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GJJ
 
 | | Multi-drug efflux pump RE-CmeB Apo form | | Descriptor: | Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GK4
 
 | | Multi-drug efflux pump RE-CmeB bound with Chloramphenicol | | Descriptor: | CHLORAMPHENICOL, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-17 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GK0
 
 | | Multi-drug efflux pump RE-CmeB bound with Erythromycin | | Descriptor: | ERYTHROMYCIN A, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-16 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
5ZSI
 
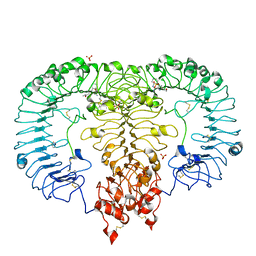 | | Crystal structure of monkey TLR7 in complex with CL097 | | Descriptor: | 2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
3OBZ
 
 | |
3OTT
 
 | |
5K07
 
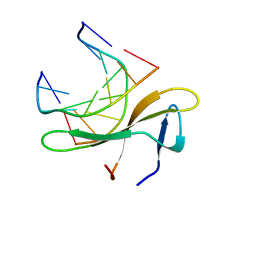 | | Crystal structure of CREN7-DSDNA (GTAATTGC) complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*GP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y. | | Deposit date: | 2016-05-17 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence-Dependent T:G Base Pair Opening in DNA Double Helix Bound by Cren7, a Chromatin Protein Conserved among Crenarchaea
PLoS ONE, 11, 2016
|
|
5K17
 
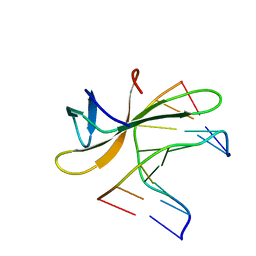 | | Crystal structure of CREN7-DSDNA (GTGATCGC) complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y. | | Deposit date: | 2016-05-17 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sequence-Dependent T:G Base Pair Opening in DNA Double Helix Bound by Cren7, a Chromatin Protein Conserved among Crenarchaea
PLoS ONE, 11, 2016
|
|
5H6V
 
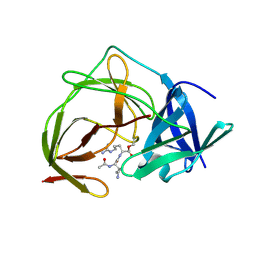 | |
8HIJ
 
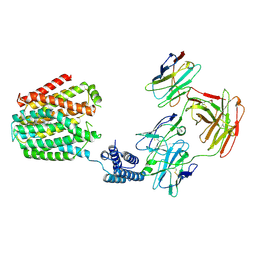 | | The 5-MTHF-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HIK
 
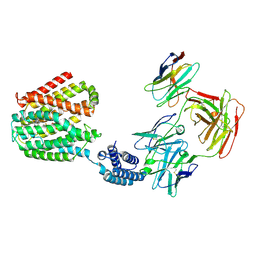 | | The TPP-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HII
 
 | | The BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | BRIL-SLC19A1 chimera, anti-BRIL Fab heavy chain, anti-BRIL Fab light chain, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
5Y28
 
 | | Crystal structure of H. pylori HtrA with PDZ2 deletion | | Descriptor: | 1,2-ETHANEDIOL, Periplasmic serine endoprotease DegP-like, UNK-UNK-UNK-UNK | | Authors: | Zhang, Z, Huang, Q, Tao, X. | | Deposit date: | 2017-07-24 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.08606815 Å) | | Cite: | The unique trimeric assembly of the virulence factor HtrA fromHelicobacter pylorioccurs via N-terminal domain swapping.
J.Biol.Chem., 294, 2019
|
|
5Y2D
 
 | | Crystal structure of H. pylori HtrA | | Descriptor: | Periplasmic serine endoprotease DegP-like, UNK-UNK-K-UNK-UNK-UNK-UNK-UNK-UNK-UNK, UNK-UNK-UNK, ... | | Authors: | Zhang, Z, Huang, Q, Tao, X. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.70009851 Å) | | Cite: | The unique trimeric assembly of the virulence factor HtrA fromHelicobacter pylorioccurs via N-terminal domain swapping.
J.Biol.Chem., 294, 2019
|
|
8HGW
 
 | | Crystal structure of MehpH in complex with MBP | | Descriptor: | 1-BUTANOL, Monoalkyl phthalate hydrolase, PHTHALIC ACID | | Authors: | Zhang, Z.M, Wang, Y.J, Chen, Y.B. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.80001163 Å) | | Cite: | Molecular insights into the catalytic mechanism of plasticizer degradation by a monoalkyl phthalate hydrolase.
Commun Chem, 6, 2023
|
|
8HGV
 
 | |
2JX0
 
 | | The paxillin-binding domain (PBD) of G Protein Coupled Receptor (GPCR)-kinase (GRK) interacting protein 1 (GIT1) | | Descriptor: | ARF GTPase-activating protein GIT1 | | Authors: | Zhang, Z, Guibao, C.D, Simmerman, J.A, Zheng, J. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | GIT1 paxillin-binding domain is a four-helix bundle, and it binds to both paxillin LD2 and LD4 motifs.
J.Biol.Chem., 283, 2008
|
|
4RDD
 
 | | Co-crystal structure of SHP2 in complex with a Cefsulodin derivative | | Descriptor: | 1-({(2R)-4-carboxy-2-[(R)-carboxy{[(2R)-2-phenyl-2-sulfoacetyl]amino}methyl]-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.Y, Yu, Z.H, He, R, Zhang, R.Y. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-01 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Exploring the Existing Drug Space for Novel pTyr Mimetic and SHP2 Inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
1JW6
 
 | | Crystal Structure of the Complex of Concanavalin A and Hexapeptide | | Descriptor: | CALCIUM ION, Concanavalin A, ISOPROPYL ALCOHOL, ... | | Authors: | Zhang, Z, Qian, M, Huang, Q, Jia, Y, Tang, Y. | | Deposit date: | 2001-09-02 | | Release date: | 2001-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the complex of concanavalin A and hexapeptide.
J.Protein Chem., 20, 2001
|
|
7WRL
 
 | |
7WRZ
 
 | | Local resolution of BD55-5840 Fab and SARS-COV2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-5840H, BD55-5840L, ... | | Authors: | Zhang, Z.Z, Xiao, J.J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
6JCJ
 
 | | Structure of crolibulin in complex with tubulin | | Descriptor: | (4R)-2,7,8-triamino-4-(3-bromo-4,5-dimethoxyphenyl)-4H-1-benzopyran-3-carbonitrile, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Yang, J. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of crolibulin in complex with tubulin provides a rationale for drug design.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
3PWT
 
 | |
6LR7
 
 | |
