5E17
 
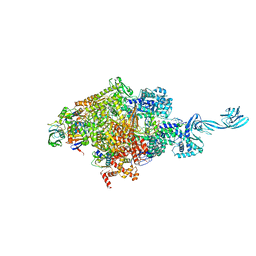 | |
5CY9
 
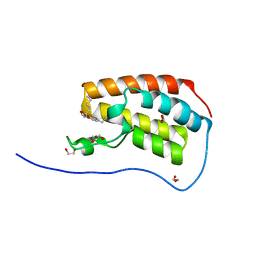 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-30 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5VTM
 
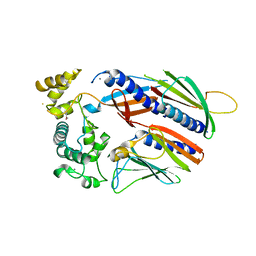 | |
5CRZ
 
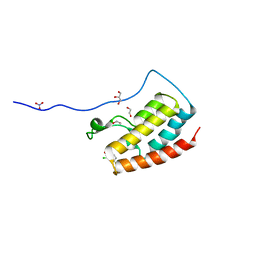 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-4-fluorobenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5D0C
 
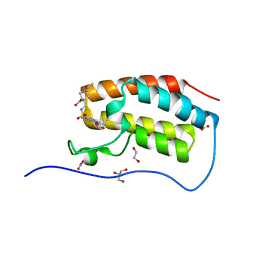 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-08-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CTL
 
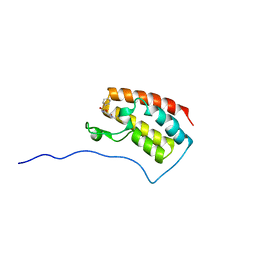 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | Bromodomain-containing protein 4, N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)benzenesulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5BW0
 
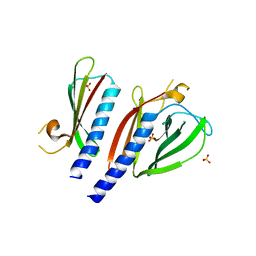 | | The crystal structure of minor pseudopilin binary complex of XcpV and XcpW from the Type 2 secretion system of Pseudomonas aeruginosa | | Descriptor: | SULFATE ION, Type II secretion system protein I, Type II secretion system protein J | | Authors: | Zhang, Y, Faucher, F, Poole, K, Jia, Z. | | Deposit date: | 2015-06-05 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided disruption of the pseudopilus tip complex inhibits the Type II secretion in Pseudomonas aeruginosa.
PLoS Pathog., 14, 2018
|
|
5D4E
 
 | |
4DC2
 
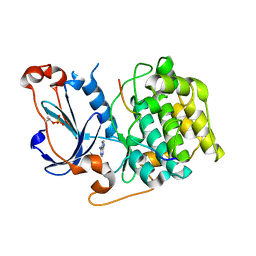 | | Structure of PKC in Complex with a Substrate Peptide from Par-3 | | Descriptor: | ADENINE, Partitioning defective 3 homolog, Protein kinase C iota type | | Authors: | Shang, Y, Wang, C, Yu, J, Zhang, M. | | Deposit date: | 2012-01-17 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKC iota in complex with a substrate peptide from Par-3
Structure, 20, 2012
|
|
5VAI
 
 | | Cryo-EM structure of the activated Glucagon-like peptide-1 receptor in complex with G protein | | Descriptor: | Glucagon-like peptide 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, Y, Sun, B, Feng, D, Hu, H, Chu, M, Qu, Q, Tarrasch, J.T, Li, S, Kobilka, T.S, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2017-03-27 | | Release date: | 2017-05-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein.
Nature, 546, 2017
|
|
5UQ7
 
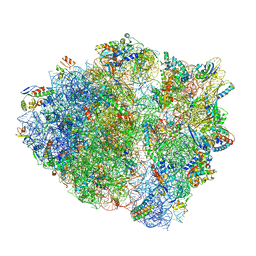 | | 70S ribosome complex with dnaX mRNA stemloop and E-site tRNA ("in" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
5V21
 
 | |
4Z69
 
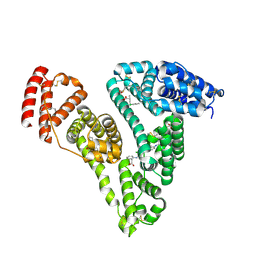 | | Human serum albumin complexed with palmitic acid and diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, PALMITIC ACID, PENTADECANOIC ACID, ... | | Authors: | Zhang, Y, Yang, F. | | Deposit date: | 2015-04-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural basis of non-steroidal anti-inflammatory drug diclofenac binding to human serum albumin.
Chem.Biol.Drug Des., 86, 2015
|
|
5D4C
 
 | |
5V22
 
 | |
5D4D
 
 | |
5UQ8
 
 | | 70S ribosome complex with dnaX mRNA stem-loop and E-site tRNA ("out" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2018-03-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
1THD
 
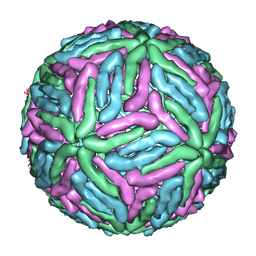 | | COMPLEX ORGANIZATION OF DENGUE VIRUS E PROTEIN AS REVEALED BY 9.5 ANGSTROM CRYO-EM RECONSTRUCTION | | Descriptor: | Major envelope protein E | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
2QCX
 
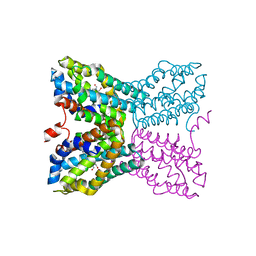 | | Crystal structure of Bacillus subtilis TenA Y112F mutant complexed with formyl aminomethyl pyrimidine | | Descriptor: | N-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-N-(2-HYDROXYETHYL)FORMAMIDE, Transcriptional activator tenA | | Authors: | Zhang, Y, Jenkins, A.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2007-06-19 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis studies on TenA: A thiamin salvage enzyme from Bacillus subtilis
Bioorg.Chem., 36, 2008
|
|
1TGE
 
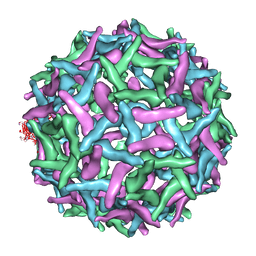 | | The structure of immature Dengue virus at 12.5 angstrom | | Descriptor: | envelope glycoprotein | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-05-28 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein.
Structure, 12, 2004
|
|
3LC8
 
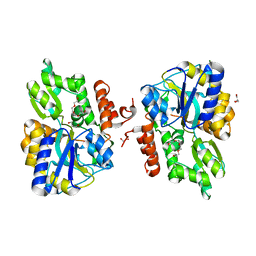 | |
2R7M
 
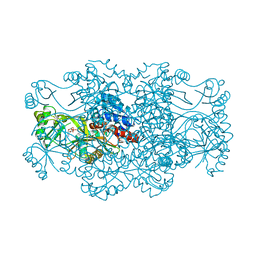 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with AMP | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R84
 
 | | Crystal structure of PurP from Pyrococcus furiosus complexed with AMP and AICAR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-10 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R7N
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ADP and FAICAR | | Descriptor: | 5-(formylamino)-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxamide, 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R7L
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ATP and AICAR | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-TRIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
