8XNG
 
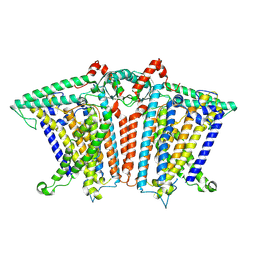 | |
8XVZ
 
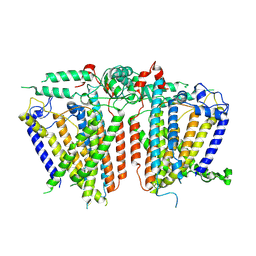 | |
8XS4
 
 | |
8XVX
 
 | |
8XW1
 
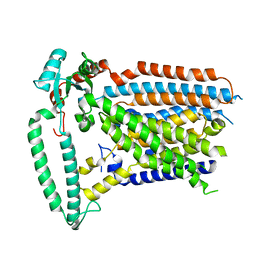 | |
8XW4
 
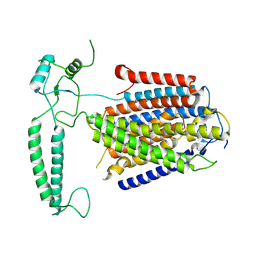 | |
8XS0
 
 | |
8XVY
 
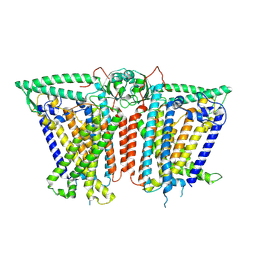 | |
8XRY
 
 | |
8XW0
 
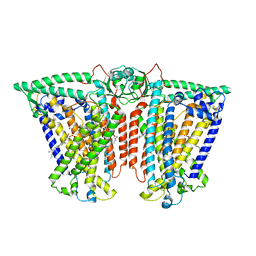 | | Cryo-EM structure of OSCA3.1-GDN state | | 分子名称: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | 著者 | Zhang, Y, Han, Y. | | 登録日 | 2024-01-15 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XS5
 
 | |
8XW2
 
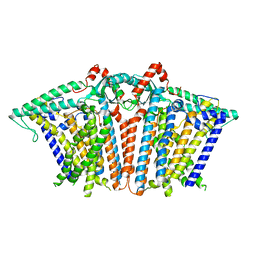 | |
8XW3
 
 | |
8XAJ
 
 | |
5W65
 
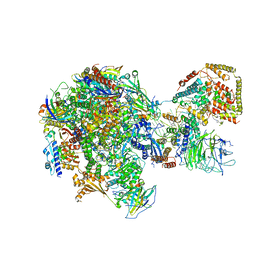 | | RNA polymerase I Initial Transcribing Complex State 2 | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2017-06-16 | | 公開日 | 2017-08-02 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W64
 
 | | RNA Polymerase I Initial Transcribing Complex State 1 | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2017-06-16 | | 公開日 | 2017-07-26 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
8WWS
 
 | | Crystal structure of cis-epoxysuccinate hydrolase from Klebsiella oxytoca with L(+)-tartaric acid | | 分子名称: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, CARBONATE ION, ... | | 著者 | Han, Y, Kong, X.D, Li, J, Xu, J.H. | | 登録日 | 2023-10-26 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural Insights of a cis -Epoxysuccinate Hydrolase Facilitate the Development of Robust Biocatalysts for the Production of l-(+)-Tartrate.
Biochemistry, 63, 2024
|
|
8WMT
 
 | |
8WNH
 
 | |
6XNY
 
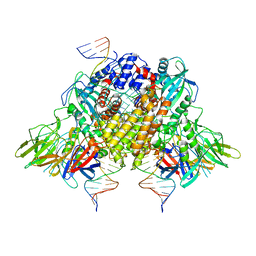 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Paired-Form) | | 分子名称: | 12RSS integration strand (55-mer), 12RSS signal DNA top strand (34-mer), 23RSS integration strand (66-mer), ... | | 著者 | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | 登録日 | 2020-07-05 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6XNZ
 
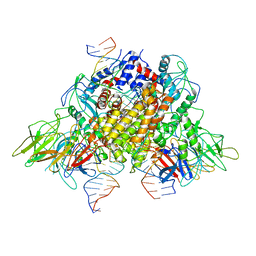 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Target Capture Complex | | 分子名称: | 12RSS integration strand (34-mer), 12RSS non-integration strand (34-mer), 23RSS integration strand (45-mer), ... | | 著者 | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | 登録日 | 2020-07-05 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | 分子名称: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | 著者 | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | 登録日 | 2015-08-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
6XGZ
 
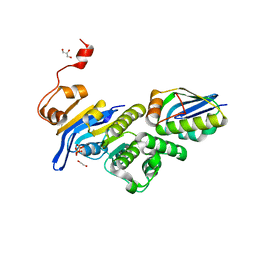 | | Crystal structure of E. coli MlaFB ABC transport subunits in the monomeric state | | 分子名称: | 1,2-ETHANEDIOL, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | 著者 | Chang, Y, Bhabha, G, Ekiert, D.C. | | 登録日 | 2020-06-18 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation.
Elife, 9, 2020
|
|
4G2V
 
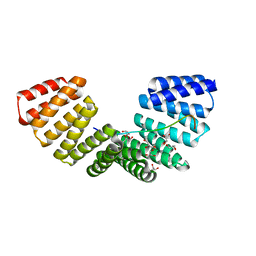 | | Structure complex of LGN binding with FRMPD1 | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, G-protein-signaling modulator 2, ... | | 著者 | Shang, Y, Pan, Z, Wen, W, Wang, W, Zhang, M. | | 登録日 | 2012-07-13 | | 公開日 | 2013-01-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and biochemical characterization of the interaction between LGN and Frmpd1
J.Mol.Biol., 425, 2013
|
|
9ATN
 
 | |
