7K65
 
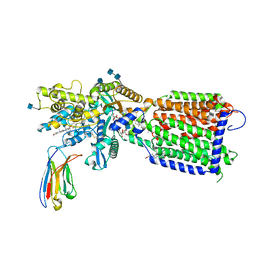 | | Hedgehog receptor Patched (PTCH1) in complex with conformation selective nanobody TI23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Zhang, Y, Bulkley, D.P, Liang, J, Manglik, A, Cheng, Y, Beachy, P.A. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hedgehog pathway activation through nanobody-mediated conformational blockade of the Patched sterol conduit.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5GKQ
 
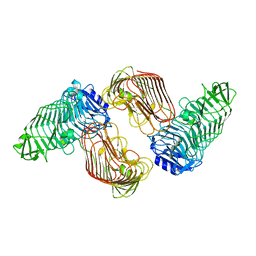 | | Structure of PL6 family alginate lyase AlyGC mutant-R241A | | Descriptor: | AlyGC mutant - R241A, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Zhang, Y.Z, Wang, P, Xu, F. | | Deposit date: | 2016-07-05 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Novel Molecular Insights into the Catalytic Mechanism of Marine Bacterial Alginate Lyase AlyGC from Polysaccharide Lyase Family 6
J. Biol. Chem., 292, 2017
|
|
5GXD
 
 | |
7DR8
 
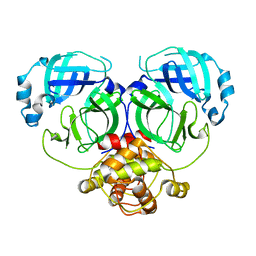 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-26 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.338149 Å) | | Cite: | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
7E7S
 
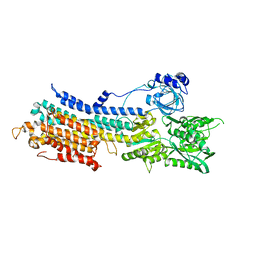 | | WT transporter state1 | | Descriptor: | CALCIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-02-27 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis provides new mechanistic insight into ATP binding to Ca 2+ -ATPase SERCA2b.
Embo J., 40, 2021
|
|
4G7O
 
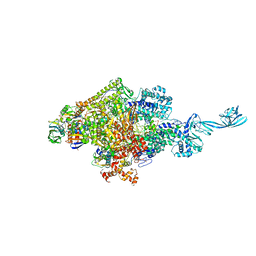 | |
4G7Z
 
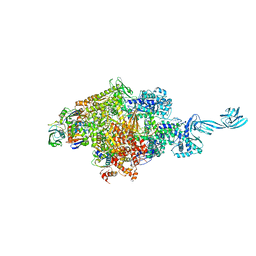 | |
8H91
 
 | |
3PWM
 
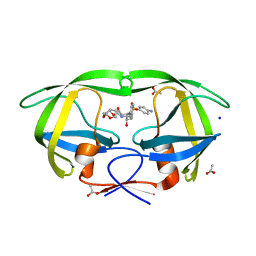 | | HIV-1 Protease Mutant L76V with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
3PY3
 
 | |
7WHM
 
 | |
7WHN
 
 | |
3PWR
 
 | | HIV-1 Protease Mutant L76V complexed with Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
7WHP
 
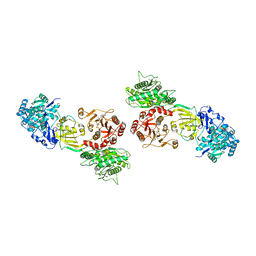 | | Pentameric turret of Bombyx mori cytoplasmic polyhedrosis virus after spike detaches. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE, Structural protein VP3 | | Authors: | Zhang, Y, Cui, Y, Sun, J, Zhou, Z.H. | | Deposit date: | 2021-12-31 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Multiple conformations of trimeric spikes visualized on a non-enveloped virus.
Nat Commun, 13, 2022
|
|
5Z1W
 
 | |
7DK8
 
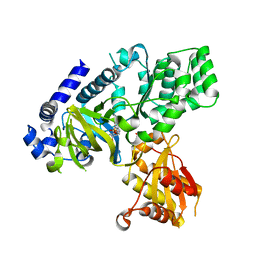 | | Crystal structure of OsGH3-8 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable indole-3-acetic acid-amido synthetase GH3.8 | | Authors: | Zhang, Y.K, Xu, G.L, Ming, Z.H. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the acyl acid amido synthetase GH3-8 from Oryza sativa.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
5D4E
 
 | |
4NPT
 
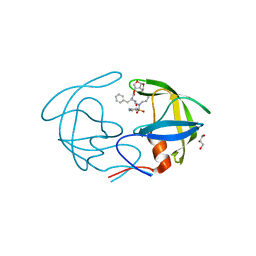 | | Crystal Structure of HIV-1 Protease Multiple Mutant P51 Complexed with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structures of darunavir-resistant HIV-1 protease mutant reveal atypical binding of darunavir to wide open flaps.
Acs Chem.Biol., 9, 2014
|
|
3PEF
 
 | |
3TG3
 
 | | Crystal structure of the MAPK binding domain of MKP7 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein phosphatase 16 | | Authors: | Zhang, Y.Y, Liu, X, Wu, J.W, Wang, Z.X. | | Deposit date: | 2011-08-17 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | A Distinct Interaction Mode Revealed by the Crystal Structure of the Kinase p38alpha with the MAPK Binding Domain of the Phosphatase MKP5.
Sci.Signal., 4, 2011
|
|
3TG1
 
 | |
6O3W
 
 | |
7D6E
 
 | |
7D6D
 
 | |
6O3Y
 
 | | Crystal structure of yeast Nrd1 CID in complex with Sen1 NIM3 | | Descriptor: | CHLORIDE ION, Helicase SEN1, Protein NRD1 | | Authors: | Zhang, Y, Tong, L. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1.
Structure, 27, 2019
|
|
