3J6C
 
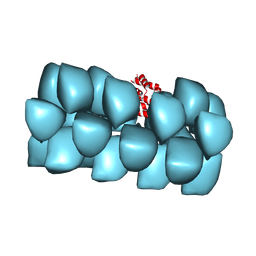 | | Cryo-EM structure of MAVS CARD filament | | 分子名称: | Mitochondrial antiviral-signaling protein | | 著者 | Xu, H, He, X, Zheng, H, Huang, L.J, Hou, F, Yu, Z, de la Cruz, M.J, Borkowski, B, Zhang, X, Chen, Z.J, Jiang, Q.-X. | | 登録日 | 2014-02-04 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.6 Å) | | 主引用文献 | Structural basis for the prion-like MAVS filaments in antiviral innate immunity.
Elife, 3, 2014
|
|
7FBI
 
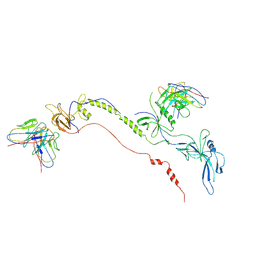 | | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5 | | 分子名称: | 3A3 heavy chain, 3A3 light chain, 3A5 heavy chain, ... | | 著者 | Zheng, Q, Li, S, Zha, Z, Hong, J, Chen, Y, Zhang, X. | | 登録日 | 2021-07-10 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5
To Be Published
|
|
7WQV
 
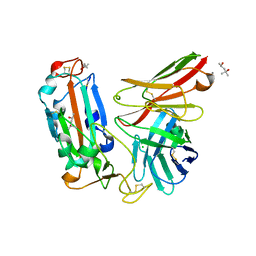 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | 著者 | Zha, J, Meng, L, Zhang, X, Li, D. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
3FDE
 
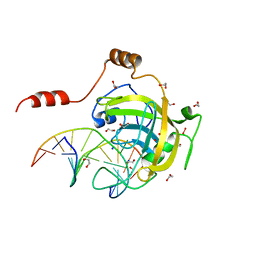 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG DNA, crystal structure in space group C222(1) at 1.4 A resolution | | 分子名称: | 1,2-ETHANEDIOL, 5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', ... | | 著者 | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | 登録日 | 2008-11-25 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
6IZJ
 
 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | 分子名称: | 1,2-ETHANEDIOL, NITRATE ION, NreA | | 著者 | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | 登録日 | 2018-12-19 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
3EA3
 
 | | Crystal Structure of the Y246S/Y247S/Y248S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | 分子名称: | 1-phosphatidylinositol phosphodiesterase, MANGANESE (II) ION | | 著者 | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | 登録日 | 2008-08-24 | | 公開日 | 2009-04-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Modulation of Bacillus thuringiensis Phosphatidylinositol-specific Phospholipase C Activity by Mutations in the Putative Dimerization Interface.
J.Biol.Chem., 284, 2009
|
|
6K2H
 
 | | structural characterization of mutated NreA protein in nitrate binding site from staphylococcus aureus. | | 分子名称: | 1,2-ETHANEDIOL, NreA | | 著者 | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | 登録日 | 2019-05-14 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
1AQL
 
 | |
6KD3
 
 | |
1VAZ
 
 | | Solution structures of the p47 SEP domain | | 分子名称: | NSFL1 cofactor p47 | | 著者 | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | 登録日 | 2004-02-20 | | 公開日 | 2004-04-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97.
Embo J., 23, 2004
|
|
6LJS
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-[(2-phenylphenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
3O06
 
 | |
1TU4
 
 | | Crystal Structure of Rab5-GDP Complex | | 分子名称: | COBALT (II) ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-5A, ... | | 著者 | Zhu, G, Zhai, P, Liu, J, Terzyan, S, Li, G, Zhang, X.C. | | 登録日 | 2004-06-24 | | 公開日 | 2004-10-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of Rab5-Rabaptin5 interaction in endocytosis
Nat.Struct.Mol.Biol., 11, 2004
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | 分子名称: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | 著者 | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2015-11-25 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
6IPO
 
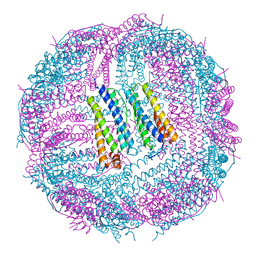 | | Ferritin mutant C90A/C102A/C130A/D144C | | 分子名称: | Ferritin heavy chain, MAGNESIUM ION | | 著者 | Zang, J, Chen, H, Zhang, X, Zhao, G. | | 登録日 | 2018-11-03 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.998 Å) | | 主引用文献 | Disulfide-mediated conversion of 8-mer bowl-like protein architecture into three different nanocages.
Nat Commun, 10, 2019
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
3O07
 
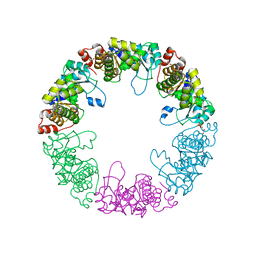 | | Crystal structure of yeast pyridoxal 5-phosphate synthase Snz1 complexed with substrate G3P | | 分子名称: | GLYCERALDEHYDE-3-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | 著者 | Teng, Y.B, Zhang, X, Hu, H.X, Zhou, C.Z. | | 登録日 | 2010-07-19 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
5FB1
 
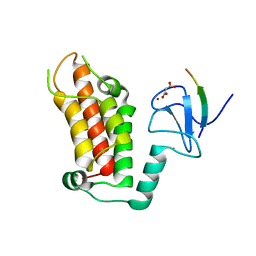 | |
1KHC
 
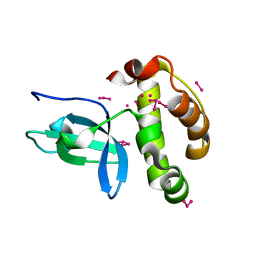 | | Crystal Structure of the PWWP Domain of Mammalian DNA Methyltransferase Dnmt3b | | 分子名称: | DNA cytosine-5 methyltransferase 3B2, UNKNOWN ATOM OR ION | | 著者 | Qiu, C, Sawada, K, Zhang, X, Cheng, X. | | 登録日 | 2001-11-29 | | 公開日 | 2002-02-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds.
Nat.Struct.Biol., 9, 2002
|
|
1AKN
 
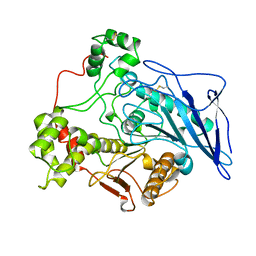 | | STRUCTURE OF BILE-SALT ACTIVATED LIPASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILE-SALT ACTIVATED LIPASE | | 著者 | Wang, X, Zhang, X. | | 登録日 | 1997-05-23 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism.
Structure, 5, 1997
|
|
1V92
 
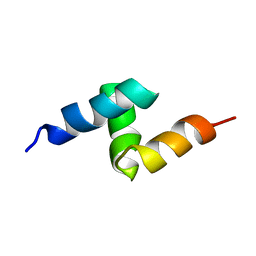 | | Solution structure of the UBA domain from p47, a major cofactor of the AAA ATPase p97 | | 分子名称: | NSFL1 cofactor p47 | | 著者 | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | 登録日 | 2004-01-19 | | 公開日 | 2004-04-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97
Embo J., 23, 2004
|
|
4HIN
 
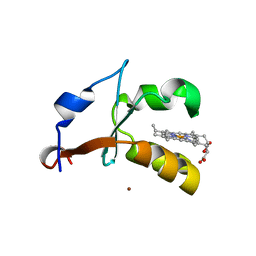 | | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L) | | 分子名称: | COPPER (II) ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Lovell, S, Battaile, K.P, Parthasarathy, S, Sun, N, Terzyan, S, Zhang, X, Rivera, M, Kuczera, K, Benson, D.R. | | 登録日 | 2012-10-11 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L)
To be Published
|
|
3EA2
 
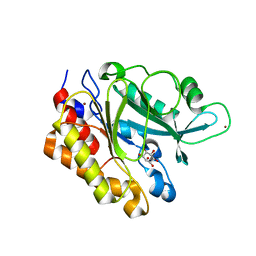 | | Crystal Structure of the Myo-inositol bound Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ZINC ION | | 著者 | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | 登録日 | 2008-08-24 | | 公開日 | 2009-04-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
4R2E
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | 著者 | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | 登録日 | 2014-08-11 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2R
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with carboxylated DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | 著者 | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | 登録日 | 2014-08-12 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.089 Å) | | 主引用文献 | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
