6GSB
 
 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Rashid, G.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, D.H. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
1F3Y
 
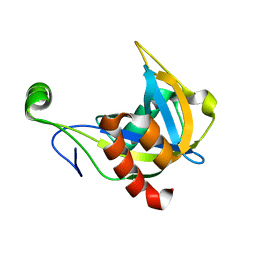 | | SOLUTION STRUCTURE OF THE NUDIX ENZYME DIADENOSINE TETRAPHOSPHATE HYDROLASE FROM LUPINUS ANGUSTIFOLIUS L. | | Descriptor: | DIADENOSINE 5',5'''-P1,P4-TETRAPHOSPHATE HYDROLASE | | Authors: | Swarbrick, J.D, Bashtannyk, T, Maksel, D, Zhang, X.R, Blackburn, G.M, Gayler, K.R, Gooley, P.R. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the Nudix enzyme diadenosine tetraphosphate hydrolase from Lupinus angustifolius L.
J.Mol.Biol., 302, 2000
|
|
6GSC
 
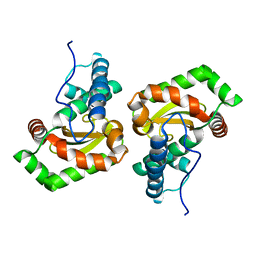 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Rashid, G.M.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, T.D.H. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
6X1A
 
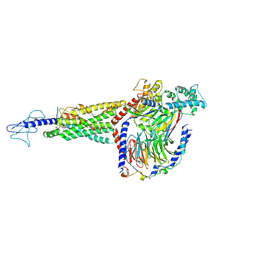 | | Non peptide agonist PF-06882961, bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
7BEG
 
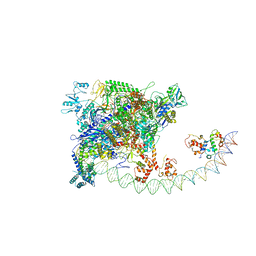 | | Structures of class I bacterial transcription complexes | | Descriptor: | Class I pacrA promoter non-template DNA, Class I pacrA promoter template DNA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Ye, F.Z, Hao, M, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
8BQW
 
 | | Cryo-EM structure of alpha-synuclein filaments doublet from Juvenile-onset synucleinopathy | | Descriptor: | Alpha-synuclein, Unknown protein | | Authors: | Yang, Y, Garringer, J.H, Shi, Y, Lovestam, S, Peak-Chew, S.Y, Zhang, X.J, Kotecha, A, Bacioglu, M, Koto, A, Takao, M, Spillantini, G.M, Ghetti, B, Vidal, R, Murzin, G.A, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2023-01-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | New SNCA mutation and structures of alpha-synuclein filaments from juvenile-onset synucleinopathy.
Acta Neuropathol, 145, 2023
|
|
8BQV
 
 | | Cryo-EM structure of alpha-synuclein singlet filament from Juvenile-onset synucleinopathy | | Descriptor: | Alpha-synuclein, Unknown protein | | Authors: | Yang, Y, Garringer, J.H, Shi, Y, Lovestam, S, Peak-Chew, S.Y, Zhang, X.J, Kotecha, A, Bacioglu, M, Koto, A, Takao, M, Spillantini, G.M, Ghetti, B, Vidal, R, Murzin, G.A, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2023-01-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | New SNCA mutation and structures of alpha-synuclein filaments from juvenile-onset synucleinopathy.
Acta Neuropathol, 145, 2023
|
|
6Z3A
 
 | | Mec1-Ddc2 (wild-type) in complex with AMP-PNP | | Descriptor: | DNA damage checkpoint protein LCD1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase MEC1, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2020-05-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6Z2X
 
 | | Mec1-Ddc2 (F2244L mutant) in complex with Mg AMP-PNP (State II) | | Descriptor: | DNA damage checkpoint protein LCD1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2020-05-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6Z2W
 
 | | Mec1-Ddc2 (F2244L mutant) in complex with Mg AMP-PNP | | Descriptor: | DNA damage checkpoint protein LCD1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2020-05-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6R9G
 
 | |
7WKK
 
 | | Cryo-EM structure of the IR subunit from X. laevis NPC | | Descriptor: | Aaas-prov protein, IL4I1 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the inner ring from the Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
4DFF
 
 | | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia | | Descriptor: | 8,9-dimethoxy-1-(1,3-thiazol-5-yl)-5,6-dihydroimidazo[5,1-a]isoquinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ho, G.D, Seganish, W.M, Bercovici, A, Tulshian, D, Greenlee, W.J, Van Rijn, R, Hruza, A, Xiao, L, Rindgen, D, Mullins, D, Guzzi, M, Zhang, X, Bleichardt, C, Hodgson, R. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DHI
 
 | | Structure of C. elegans OTUB1 bound to human UBC13 | | Descriptor: | Ubiquitin thioesterase otubain-like, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
2QJO
 
 | | crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase (NadM) complexed with ADPRP and NAD from Synechocystis sp. | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Bifunctional NMN adenylyltransferase/Nudix hydrolase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-08 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
2R5W
 
 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
4DHZ
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBC13~Ub | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | Deposit date: | 2015-09-05 | | Release date: | 2015-10-21 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
2KA9
 
 | | Solution structure of PSD-95 PDZ12 complexed with cypin peptide | | Descriptor: | Disks large homolog 4, cypin peptide | | Authors: | Wang, W.N, Weng, J.W, Zhang, X, Liu, M.L, Zhang, M.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95
J.Am.Chem.Soc., 131, 2009
|
|
1P9E
 
 | | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3 | | Descriptor: | CADMIUM ION, Methyl Parathion Hydrolase, POTASSIUM ION, ... | | Authors: | Dong, Y, Sun, L, Bartlam, M, Rao, Z, Zhang, X. | | Deposit date: | 2003-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3
To be Published
|
|
2K9X
 
 | | Solution structure of Urm1 from Trypanosoma brucei | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, W, Zhang, J, Xu, C, Wang, T, Zhang, X, Tu, X. | | Deposit date: | 2008-10-27 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 from Trypanosoma brucei
Proteins, 75, 2009
|
|
3TC1
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori | | Descriptor: | MAGNESIUM ION, Octaprenyl Pyrophosphate Synthase | | Authors: | Zhang, J.Y, Zhang, X.L, Li, D.F, Zou, Q.M, Wang, D.C. | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori
To be Published
|
|
1N6H
 
 | | Crystal Structure of Human Rab5a | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Zhu, G, Liu, J, Terzyan, S, Zhai, P, Li, G, Zhang, X.C. | | Deposit date: | 2002-11-11 | | Release date: | 2002-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | High Resolution Crystal Structures of Human Rab5a and Five Mutants with Substitutions in the Catalytically Important Phosphate-Binding Loop
J.Biol.Chem., 278, 2003
|
|
1N6I
 
 | | Crystal Structure of Human Rab5a A30P mutant Complex with GDP | | Descriptor: | BETA-MERCAPTOETHANOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhu, G, Liu, J, Terzyan, S, Zhai, P, Li, G, Zhang, X.C. | | Deposit date: | 2002-11-11 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High Resolution Crystal Structures of Human Rab5a and Five Mutants with Substitutions in the Catalytically Important Phosphate-Binding Loop
J.Biol.Chem., 278, 2003
|
|
1N6K
 
 | | Crystal Structure of Human Rab5a A30P mutant complex with GDP and aluminum fluoride | | Descriptor: | ALUMINUM FLUORIDE, BETA-MERCAPTOETHANOL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhu, G, Liu, J, Terzyan, S, Zhai, P, Li, G, Zhang, X.C. | | Deposit date: | 2002-11-11 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High Resolution Crystal Structures of Human Rab5a and Five Mutants with Substitutions in the Catalytically Important Phosphate-Binding Loop
J.Biol.Chem., 278, 2003
|
|
