4V6E
 
 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4V6D
 
 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.814 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4MHA
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1817 | | Descriptor: | 2-(butylamino)-4-[(trans-4-hydroxycyclohexyl)amino]-N-(4-sulfamoylbenzyl)pyrimidine-5-carboxamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, W, Mciver, A, Stashko, M.A, Deryckere, D, Branchford, B.R, Hunter, D, Kireev, D.B, Miley, D.B.M, Norris-Drouin, J, Stewart, W.M, Lee, M, Sather, S, Zhou, Y, Dipaola, J.A, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Discovery of Mer specific tyrosine kinase inhibitors for the treatment and prevention of thrombosis.
J.Med.Chem., 56, 2013
|
|
5WCO
 
 | | Matrix Protein (M1) of Infectious Salmon Anaemia Virus | | Descriptor: | NS2 | | Authors: | Zhang, W, Zheng, W, Toh, Y, Betancourt, M.A, Tu, J, Fan, Y, Vakharia, V, Liu, J, McNew, J.A, Jin, M, Tao, Y.J. | | Deposit date: | 2017-07-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of an orthomyxovirus matrix protein reveals mechanisms for self-polymerization and membrane association.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6U6J
 
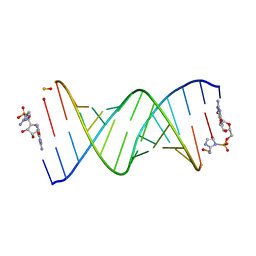 | | RNA-monomer complex containing pyrophosphate linkage | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(hydroxy)phosphoryl]guanosine, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(DPG))-3') | | Authors: | Zhang, W, Szostak, J.W, Giurgiu, C, Wright, T. | | Deposit date: | 2019-08-29 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prebiotically Plausible "Patching" of RNA Backbone Cleavage through a 3'-5' Pyrophosphate Linkage.
J.Am.Chem.Soc., 141, 2019
|
|
6L13
 
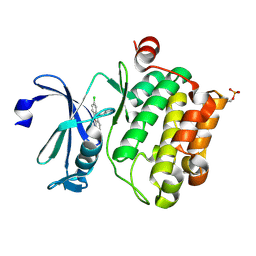 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-(2-piperidin-4-ylethyl)phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L14
 
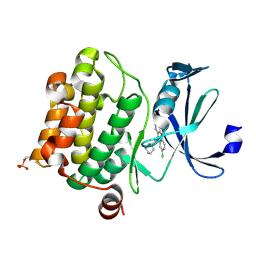 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-[3-[(3~{S})-piperidin-3-yl]propyl]phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
5ZOF
 
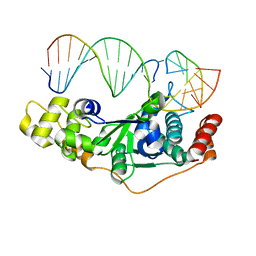 | | Crystal Structure of D181A/R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5ZOD
 
 | | Crystal Structure of hFen1 in apo form | | Descriptor: | Flap endonuclease 1, MAGNESIUM ION, POTASSIUM ION | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5ZOG
 
 | | Crystal Structure of R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5ZOE
 
 | | Crystal Structure of D181A hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5H7E
 
 | | Crystal Structure of native drCPDase | | Descriptor: | GLYCEROL, RNA 2',3'-cyclic phosphodiesterase, SULFATE ION | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the RNA 2',3'-cyclic phosphodiesterase from Deinococcus radiodurans
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5H7F
 
 | |
4RHD
 
 | | DNA Duplex with Novel ZP Base Pair | | Descriptor: | DNA 9mer novel P nucleobase, DNA 9mer novel Z nucleobase, MAGNESIUM ION | | Authors: | Zhang, W, Zhang, L, Benner, S, Huang, Z. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of functional six-nucleotide DNA.
J.Am.Chem.Soc., 137, 2015
|
|
4WW5
 
 | | Crystal structure of binary complex Bud32-Cgi121 in complex with AMPP | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, ... | | Authors: | Zhang, W. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WW9
 
 | |
4X3X
 
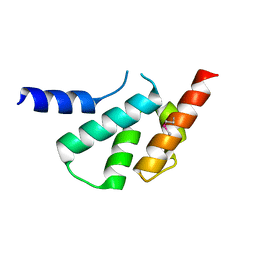 | | The crystal structure of Arc C-lobe | | Descriptor: | Activity-regulated cytoskeleton-associated protein | | Authors: | Zhang, W, Ward, M, Leahy, D, Worley, P. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
4WQ5
 
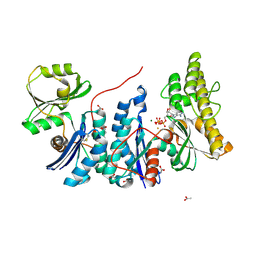 | | YgjD(V85E)-YeaZ heterodimer in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, FE (III) ION, ... | | Authors: | Zhang, W. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
4WWA
 
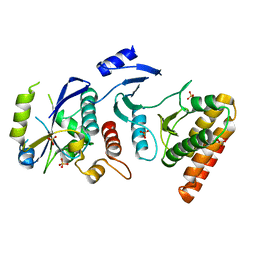 | | Crystal structure of binary complex Bud32-Cgi121 | | Descriptor: | EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, SULFATE ION | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WQ4
 
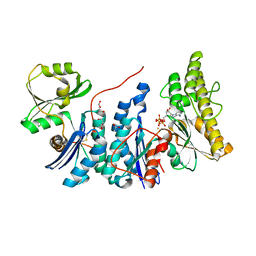 | | E. coli YgjD(E12A)-YeaZ heterodimer in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, W, Collinet, B. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
4WW7
 
 | |
4WX8
 
 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1 | | Authors: | Zhang, W, Van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WXA
 
 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1, ... | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4X3H
 
 | | CRYSTAL STRUCTURE OF ARC N-LOBE COMPLEXED WITH STARGAZIN PEPTIDE | | Descriptor: | Activity-regulated cytoskeleton-associated protein, VOLTAGE-DEPENDENT CALCIUM CHANNEL GAMMA-2 SUBUNIT | | Authors: | zhang, W, ward, m, leahy, d, worley, p. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
4X3I
 
 | | The crystal structure of Arc N-lobe complexed with CAMK2A fragment | | Descriptor: | Activity-regulated cytoskeleton-associated protein, CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II SUBUNIT ALPHA | | Authors: | Zhang, W, Ward, M, Leahy, D, Worley, P. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
