8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3Z
 
 | |
7YEO
 
 | |
8HAS
 
 | | NARROW LEAF 1-close from Japonica | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein NARROW LEAF 1 | | Authors: | Zhang, S.J, He, Y.J, Wang, N, Zhang, W.J, Liu, C.M. | | Deposit date: | 2022-10-26 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | NARROW LEAF 1-close from Japonica
To Be Published
|
|
8HAT
 
 | |
8HAU
 
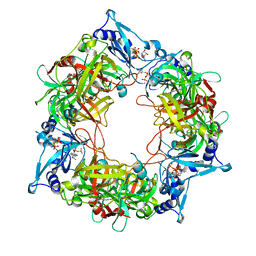 | | NARROW LEAF 1 from Indica | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein NARROW LEAF 1 | | Authors: | Zhang, S.J, He, Y.J, Wang, N, Zhang, W.J, Liu, C.M. | | Deposit date: | 2022-10-26 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | NARROW LEAF 1 from Indica
To Be Published
|
|
6IGN
 
 | |
6IGV
 
 | |
7WX7
 
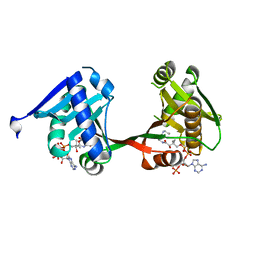 | | complex of a legionella acetyltransferase VipF and COA/ACO | | Descriptor: | ACETYL COENZYME *A, COENZYME A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Zhang, S.J, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WX5
 
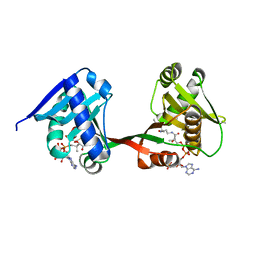 | | a Legionella acetyltransferase effector VipF | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Zhang, S.J, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | Descriptor: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y2W
 
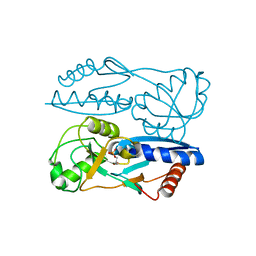 | | Structure of Synechocystis PCC6803 CcmR regulatory domain in complex with 2-PG | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Cao, D.D, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7D6C
 
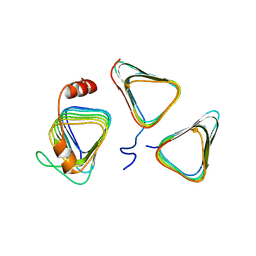 | | Crystal structure of CcmM N-terminal domain in complex with CcmN | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Carboxysome assembly protein CcmN | | Authors: | Sun, H, Cui, N, Han, S.J, Chen, Z.P, Xia, L.Y, Chen, Y, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Complex structure reveals CcmM and CcmN form a heterotrimeric adaptor in beta-carboxysome.
Protein Sci., 30, 2021
|
|
6N8A
 
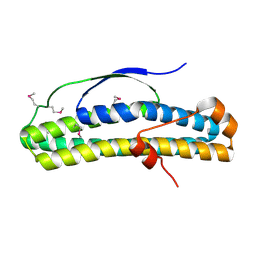 | | Crystal structure of selenomethionine-containing AcaB from uropathogenic E. coli | | Descriptor: | CHLORIDE ION, transcription regulator AcaB | | Authors: | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | Deposit date: | 2018-11-28 | | Release date: | 2020-07-15 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (3.4011 Å) | | Cite: | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
6N8B
 
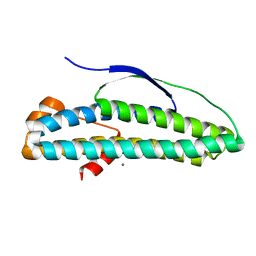 | | Crystal structure of transcription regulator AcaB from uropathogenic E. coli | | Descriptor: | CALCIUM ION, transcription regulator AcaB | | Authors: | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | Deposit date: | 2018-11-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
6HN5
 
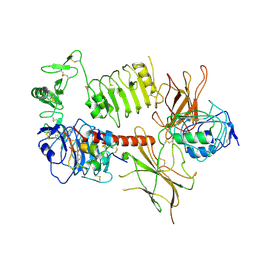 | | Leucine-zippered human insulin receptor ectodomain with single bound insulin - "upper" membrane-distal part | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Weis, F, Menting, J.G, Margetts, M.B, Chan, S.J, Xu, Y, Tennagels, N, Wohlfart, P, Langer, T, Mueller, C.W, Dreyer, M.K, Lawrence, M.C. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The signalling conformation of the insulin receptor ectodomain.
Nat Commun, 9, 2018
|
|
2ETL
 
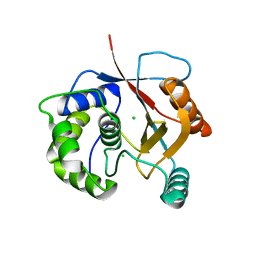 | | Crystal Structure of Ubiquitin Carboxy-terminal Hydrolase L1 (UCH-L1) | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Das, C, Hoang, Q.Q, Kreinbring, C.A, Luchansky, S.J, Meray, R.K, Ray, S.S, Lansbury, P.T, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-10-27 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
6IHG
 
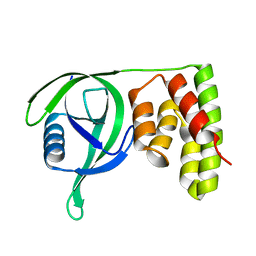 | | N terminal domain of Mycobacterium avium complex Lon protease | | Descriptor: | Lon protease | | Authors: | Chen, X.Y, Zhang, S.J, Bi, F.K, Guo, C.Y, Yao, H.W, Lin, D.H. | | Deposit date: | 2018-09-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
6HN4
 
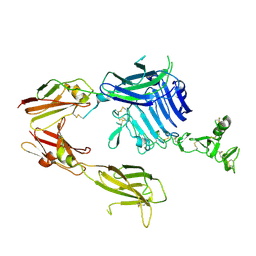 | | Leucine-zippered human insulin receptor ectodomain with single bound insulin - "lower" membrane-proximal part | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin receptor,Insulin receptor,General control protein GCN4 | | Authors: | Weis, F, Menting, J.G, Margetts, M.B, Chan, S.J, Xu, Y, Tennagels, N, Wohlfart, P, Langer, T, Mueller, C.W, Dreyer, M.K, Lawrence, M.C. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The signalling conformation of the insulin receptor ectodomain.
Nat Commun, 9, 2018
|
|
2OPP
 
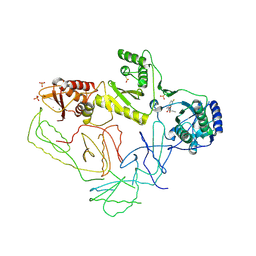 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Chan, S.J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
