6LZ3
 
 | |
7WBQ
 
 | |
7WBP
 
 | |
7WSK
 
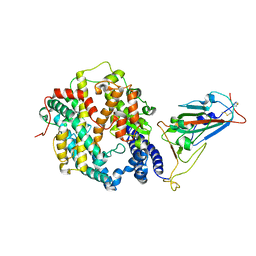 | | Crystal structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with civet ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Huang, B, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
8Y7E
 
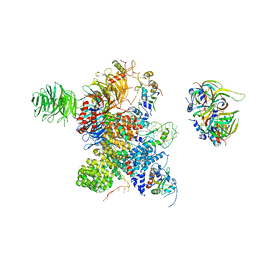 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U12 snRNP part | | Descriptor: | PHD finger-like domain-containing protein 5A, Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
8Y6O
 
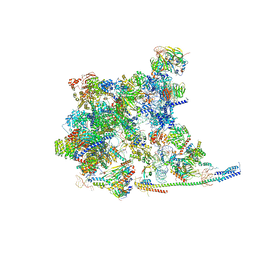 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U11 and tri-snRNP part | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Centrosomal AT-AC splicing factor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
7QHD
 
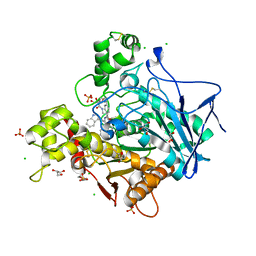 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((2-(1H-indol-3-yl)ethyl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-[2-(1~{H}-indol-3-yl)ethylcarbamoyl]phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
7QHE
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((naphthalen-1-yl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-(naphthalen-1-ylcarbamoyl)phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
1QCO
 
 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH FUMARATE AND ACETOACETATE | | Descriptor: | ACETOACETIC ACID, CALCIUM ION, FUMARIC ACID, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-17 | | Release date: | 2000-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
7W6R
 
 | | Structure of Bat coronavirus RaTG13 spike receptor-binding domain complexed with its receptor equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2021-12-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses
Nat Commun, 13, 2022
|
|
7W6U
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain complexed with its receptor equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2021-12-02 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses
Nat Commun, 13, 2022
|
|
7WSF
 
 | |
7WSE
 
 | |
7WSH
 
 | | Cryo-EM structure of SARS-CoV-2 spike receptor-binding domain in complex with sea lion ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, S, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cross-species recognition and molecular basis of SARS-CoV-2 and SARS-CoV binding to ACE2s of marine animals.
Natl Sci Rev, 9, 2022
|
|
7WSG
 
 | |
7XBY
 
 | | The crystal structure of SARS-CoV-2 Omicron BA.1 variant RBD in complex with equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, BROMIDE ION, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses.
Nat Commun, 13, 2022
|
|
6LPW
 
 | |
3VM7
 
 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | Deposit date: | 2011-12-09 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
8IYM
 
 | | Crystal structure of a protein acetyltransferase, HP0935 | | Descriptor: | 1,2-ETHANEDIOL, N-acetyltransferase domain-containing protein, POTASSIUM ION, ... | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935
To be published
|
|
8IYO
 
 | | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase domain-containing protein | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form
To be published
|
|
9B3G
 
 | | Human Notch-1 EGFs 21-23 | | Descriptor: | BARIUM ION, Neurogenic locus notch homolog protein 1 | | Authors: | Johnson, S, Sheppard, D, Handford, P.A, Lea, S.M. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and functional studies of the EGF20-27 region reveal new features of the human Notch receptor important for optimal activation
To Be Published
|
|
9B3N
 
 | | Human Notch-1 EGFs 20-24 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Neurogenic locus notch homolog protein 1, ... | | Authors: | Johnson, S, Suckling, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional studies of the EGF20-27 region reveal new features of the human Notch receptor important for optimal activation
To Be Published
|
|
4XHR
 
 | | Structure of a phospholipid trafficking complex, native | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Yu, F, He, F, Wang, C, Zhang, P. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
4XIZ
 
 | | Structure of a phospholipid trafficking complex with substrate | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Yu, F, He, F, Wang, C, Zhang, P. | | Deposit date: | 2015-01-08 | | Release date: | 2015-07-01 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
1UUB
 
 | |
