6KWK
 
 | | Crystal structure of pSLA-1*0401 complex with FMDV-derived epitope MTAHITVPY | | 分子名称: | Beta-2-microglobulin, MHC class I antigen, peptide | | 著者 | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | 登録日 | 2019-09-07 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
7TJ0
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor SL-4-241 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ACETATE ION | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
6LF8
 
 | | Crystal structure of pSLA-1*0401 complex with dodecapeptide RVEDVTNTAEYW | | 分子名称: | ARG-VAL-GLU-ASP-VAL-THR-ASN-THR-ALA-GLU-TYR-TRP, Beta-2-microglobulin, MHC class I antigen | | 著者 | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | 登録日 | 2019-11-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and Peptidomes of Swine MHC Class I with Long Peptides Reveal the Cross-Species Characteristics of the Novel N-Terminal Extension Presentation Mode.
J Immunol., 208, 2022
|
|
7C1H
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA | | 分子名称: | Non-ribosomal peptide synthetase modules | | 著者 | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | 登録日 | 2020-05-04 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1R
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | 著者 | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | 登録日 | 2020-05-05 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1P
 
 | | Crystal structure of the starter condensation domain of the rhizomide synthetase RzmA mutant H140V, R148A | | 分子名称: | Non-ribosomal peptide synthetase modules | | 著者 | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D. | | 登録日 | 2020-05-05 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1S
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA and Leu-SNAC | | 分子名称: | Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A, S-(2-acetamidoethyl) (2S)-2-azanyl-4-methyl-pentanethioate | | 著者 | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | 登録日 | 2020-05-05 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.586 Å) | | 主引用文献 | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
8PYH
 
 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Crinalium epipsammum PCC 9333 | | 分子名称: | ACETATE ION, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | 著者 | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | 登録日 | 2023-07-25 | | 公開日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
8PZK
 
 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Gloeocapsa sp. PCC 7428 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, GLYCEROL, ... | | 著者 | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | 登録日 | 2023-07-27 | | 公開日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
7CPO
 
 | | Crystal Structure of Anolis carolinensis MHC I complex | | 分子名称: | HIS-VAL-TYR-GLY-PRO-LEU-LYS-PRO-ILE, MHC CLASS I ANTIGEN, beta2-microglobulin | | 著者 | Wang, Y, Qu, Z, Ma, L, Wei, X, Zhang, N, Xia, C. | | 登録日 | 2020-08-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Crystal Structure of the MHC Class I (MHC-I) Molecule in the Green Anole Lizard Demonstrates the Unique MHC-I System in Reptiles.
J Immunol., 206, 2021
|
|
6KNM
 
 | | Apelin receptor in complex with single domain antibody | | 分子名称: | Apelin receptor,Rubredoxin,Apelin receptor, Single domain antibody JN241, ZINC ION | | 著者 | Ma, Y.B, Ding, Y, Song, X, Ma, X, Li, X, Zhang, N, Song, Y, Sun, Y, Shen, Y, Zhong, W, Hu, L.A, Ma, Y.L, Zhang, M.Y. | | 登録日 | 2019-08-06 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure-guided discovery of a single-domain antibody agonist against human apelin receptor.
Sci Adv, 6, 2020
|
|
1KJ1
 
 | | MANNOSE-SPECIFIC AGGLUTININ (LECTIN) FROM GARLIC (ALLIUM SATIVUM) BULBS COMPLEXED WITH ALPHA-D-MANNOSE | | 分子名称: | alpha-D-mannopyranose, lectin I, lectin II | | 著者 | Ramachandraiah, G, Chandra, N.R, Surolia, A, Vijayan, M. | | 登録日 | 2001-12-04 | | 公開日 | 2002-02-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Re-refinement using reprocessed data to improve the quality of the structure: a case study involving garlic lectin.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5YLX
 
 | | Integrated illustration of a valid epitope based on the SLA class I structure and tetramer technique could carry forward the development of molecular vaccine in swine species | | 分子名称: | Beta-2-microglobulin, MHC class I antigen, PRRSV-NSP9-TMP9 peptide | | 著者 | Pan, X.C, Wei, X.H, Zhang, N, Xia, C. | | 登録日 | 2017-10-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2020-05-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Illumination of PRRSV Cytotoxic T Lymphocyte Epitopes by the Three-Dimensional Structure and Peptidome of Swine Lymphocyte Antigen Class I (SLA-I).
Front Immunol, 10, 2019
|
|
7CSQ
 
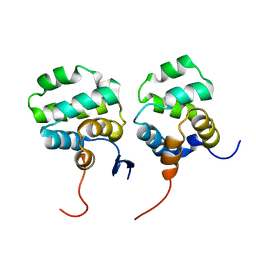 | | Solution structure of the complex between p75NTR-DD and TRADD-DD | | 分子名称: | Tumor necrosis factor receptor superfamily member 16, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | 著者 | Lin, Z, Zhang, N. | | 登録日 | 2020-08-16 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of NF-kappa B signaling by the p75 neurotrophin receptor interaction with adaptor protein TRADD through their respective death domains.
J.Biol.Chem., 297, 2021
|
|
6KCZ
 
 | |
7JLF
 
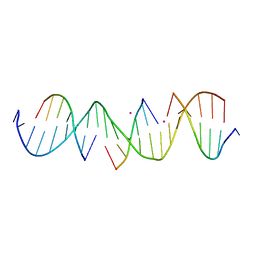 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J24 immobile Holliday junction with R3 symmetry | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*AP*GP*AP*CP*GP*TP*CP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*G)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-29 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.909 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JHA
 
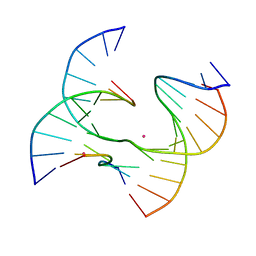 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J23 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-20 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.107 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JHV
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J36 immobile Holliday junction with R3 symmetry | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-21 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.058 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JI7
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J33 immobile Holliday junction with R3 symmetry | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*TP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*CP*AP*TP*GP*TP*CP*GP*T)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-22 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.105 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKE
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J2 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*AP*CP*GP*AP*GP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-28 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.068 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JI6
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J34 immobile Holliday junction with R3 symmetry | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*GP*TP*TP*GP*TP*CP*GP*T)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-22 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.005 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JLD
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J22 immobile Holliday junction with R3 symmetry | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*AP*CP*GP*GP*CP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*G)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-29 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.163 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKD
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J1 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*AP*CP*GP*GP*AP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*T)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-28 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.058 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JLA
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J16 immobile Holliday junction with R3 symmetry | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*GP*TP*GP*AP*CP*GP*CP*CP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*G)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-29 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JFW
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J10 immobile Holliday junction | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*CP*AP*CP*TP*CP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.012 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
