6DA6
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes, apo form at 2.6 A resolution (P212121) | | Descriptor: | GLYCEROL, MAGNESIUM ION, UNKNOWN LIGAND, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA9
 
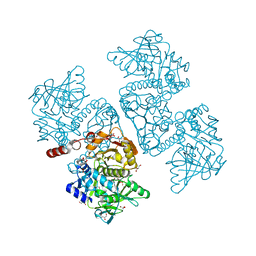 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6TGB
 
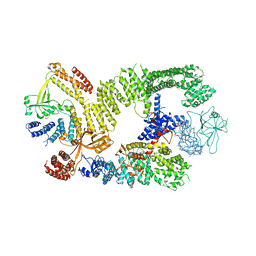 | | CryoEM structure of the binary DOCK2-ELMO1 complex | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
5HOQ
 
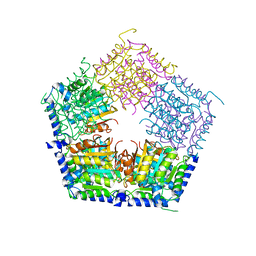 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
6C92
 
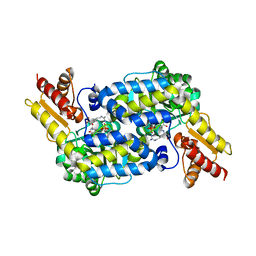 | | The structure of MppP soaked with the product 2-ketoarginine | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
6C9B
 
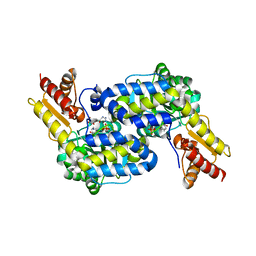 | | The structure of MppP soaked with the products 4HKA and 2KA | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-26 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
6C8T
 
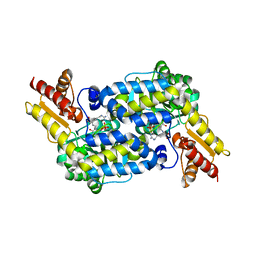 | | The structure of MppP soaked with the substrate L-Arg | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
4UI9
 
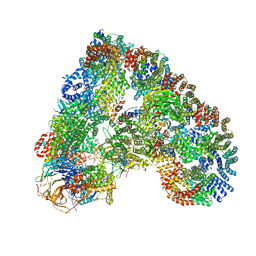 | | Atomic structure of the human Anaphase-Promoting Complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
6OMV
 
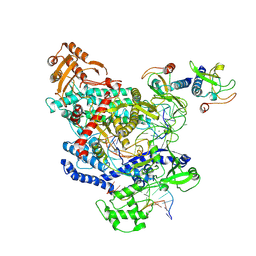 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4-DNA complex | | Descriptor: | AcrVA4, Cpf1, DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*AP*GP*GP*A)-3'), ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-04-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6XMF
 
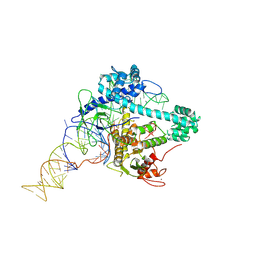 | |
6NMD
 
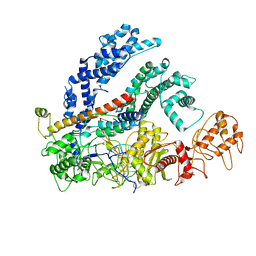 | | cryo-EM Structure of the LbCas12a-crRNA-AcrVA1 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NMA
 
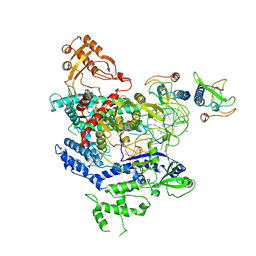 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NM9
 
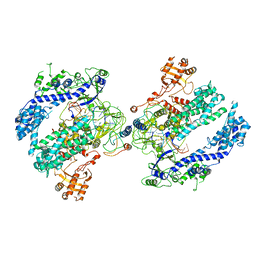 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4 dimer | | Descriptor: | AcrVA4, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NME
 
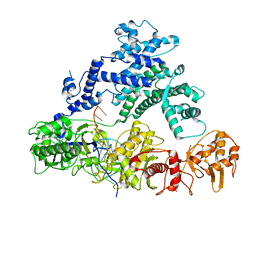 | | Structure of LbCas12a-crRNA | | Descriptor: | Cpf1, MAGNESIUM ION, crRNA | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.67 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NMC
 
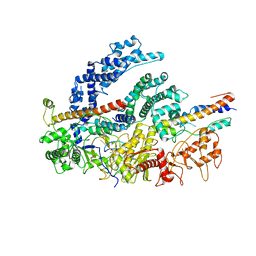 | | CryoEM structure of the LbCas12a-crRNA-2xAcrVA1 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6XMG
 
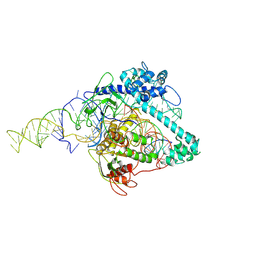 | | Cryo-EM structure of Cas12g ternary complex | | Descriptor: | CRISPR-Cas, RNA (130-MER), RNA (5'-R(P*UP*UP*AP*AP*UP*GP*CP*GP*GP*UP*AP*GP*UP*UP*UP*AP*UP*CP*AP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-06-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of the RNA-guided ribonuclease Cas12g.
Nat.Chem.Biol., 17, 2021
|
|
7OQ6
 
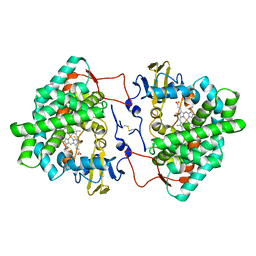 | | Crystal structure of cytochrome P450 Sas16 from Streptomyces asterosporus | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Zhang, L, Zhang, S, Bechthold, A, Einsle, O. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P450-mediated dehydrotyrosine formation during WS9326 biosynthesis proceeds via dehydrogenation of a specific acylated dipeptide substrate.
Acta Pharm Sin B, 13, 2023
|
|
6UVN
 
 | | CryoEM structure of VcCascasde-TniQ complex | | Descriptor: | Cas6, Cas7, Cas8/5, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a type I-F CRISPR RNA guided surveillance complex bound to transposition protein TniQ.
Cell Res., 30, 2020
|
|
6W62
 
 | | Cryo-EM structure of Cas12i-crRNA complex | | Descriptor: | Cas12i, crRNA | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7QUW
 
 | | CVB3-3Cpro in complex with inhibitor MG-78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, Protease 3C | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
6W5C
 
 | | Cryo-EM structure of Cas12i(E894A)-crRNA-dsDNA complex | | Descriptor: | Cas12i, NTS, Substrate, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W64
 
 | |
5A31
 
 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
