5AQO
 
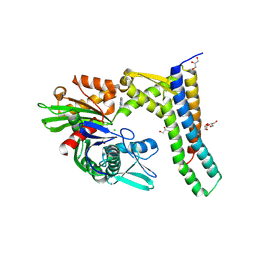 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-METHYLQUINAZOLIN-4-AMINE, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQG
 
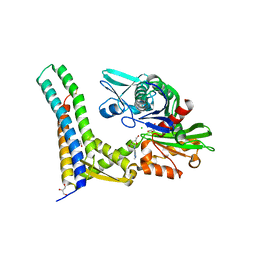 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | (2R,3R,4S,5R)-2-(3-AMINO-5-METHYL-1,4,5,6,8-PENTAAZAACENAPHTHYLEN-1(5H)-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
6KZW
 
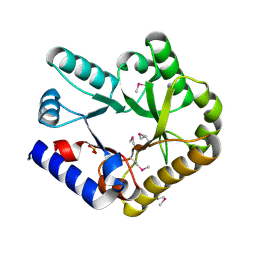 | | Crystal structure of YggS family pyridoxal phosphate-dependent enzyme PipY from Fusobacterium nucleatum | | Descriptor: | PHOSPHATE ION, Pyridoxal phosphate homeostasis protein | | Authors: | Chen, Y, Wang, L, Shang, F, Lan, J, Liu, W, Xu, Y. | | Deposit date: | 2019-09-25 | | Release date: | 2019-10-16 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of YggS family pyridoxal phosphate-dependent enzyme PipY from Fusobacterium nucleatum
To Be Published
|
|
6L3E
 
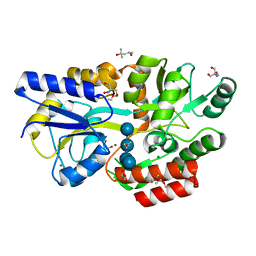 | | Crystal structure of Salmonella enterica sugar-binding protein MalE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Salmonella enterica sugar-binding protein MalE
To Be Published
|
|
6L0Z
 
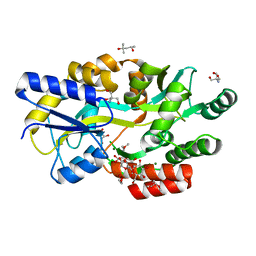 | | The crystal structure of Salmonella enterica sugar-binding protein MalE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-anhydro-D-glucitol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Salmonella enterica sugar-binding protein MalE
To Be Published
|
|
6E8W
 
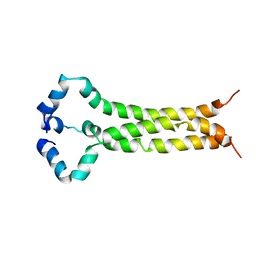 | | MPER-TM Domain of HIV-1 envelope glycoprotein (Env) | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Fu, Q, Shaik, M.M, Cai, Y, Ghantous, F, Piai, A, Peng, H, Rits-Volloch, S, Liu, Z, Harrison, S.C, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane proximal external region of HIV-1 envelope glycoprotein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6L19
 
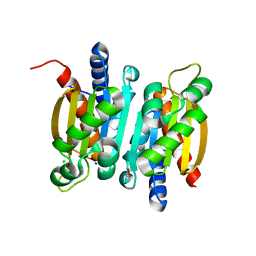 | | The crystal structure of competence or damage-inducible protein from Enterobacter asburiae | | Descriptor: | CHLORIDE ION, GLYCEROL, PncC family amidohydrolase, ... | | Authors: | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | Deposit date: | 2019-09-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The crystal structure of Competence or damage-inducible protein from Enterobacter asburiae
To Be Published
|
|
4I5I
 
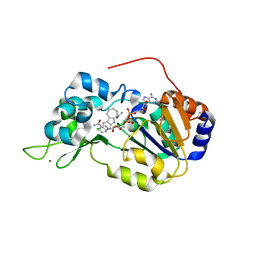 | | Crystal structure of the SIRT1 catalytic domain bound to NAD and an EX527 analog | | Descriptor: | (6S)-2-chloro-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhao, X, Allison, D, Condon, B, Zhang, F, Gheyi, T, Zhang, A, Ashok, S, Russell, M, Macewan, I, Qian, Y, Jamison, J.A, Luz, J.G. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 angstrom crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition.
J.Med.Chem., 56, 2013
|
|
4HXX
 
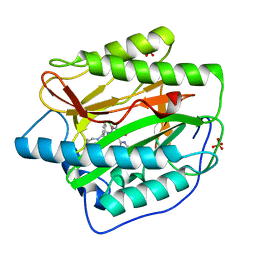 | | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1 | | Descriptor: | (1R)-N~2~-[5-chloro-2-(5-chloropyridin-2-yl)-6-methylpyrimidin-4-yl]-1-phenyl-N~1~-(4-phenylbutyl)ethane-1,2-diamine, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Liu, J, Amzel, L.M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1.
Bioorg.Med.Chem., 21, 2013
|
|
6JFI
 
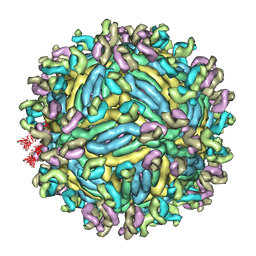 | | The symmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural protein E, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JFH
 
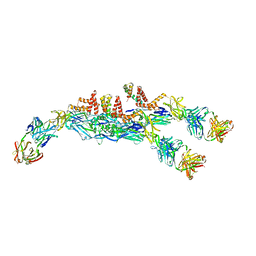 | | The asymmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural E protein, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JEP
 
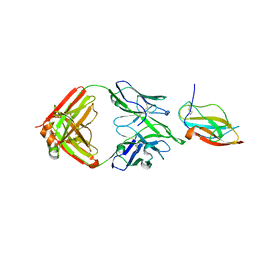 | | Structure of a neutralizing antibody bound to the Zika envelope protein domain III | | Descriptor: | Genome polyprotein, heavy chain of Fab ZK2B10, light chain of Fab ZK2B10 | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-07 | | Release date: | 2019-05-15 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
8UYO
 
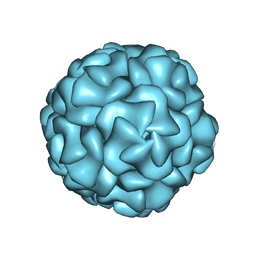 | |
7LPE
 
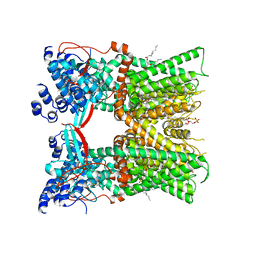 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 48 degrees Celsius, in an open state, class 1 | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LPA
 
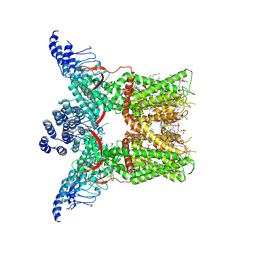 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 4 degrees Celsius | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LP9
 
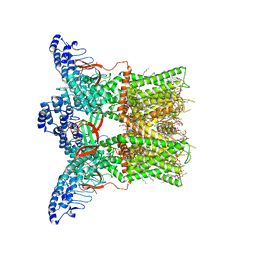 | | Cryo-EM structure of full-length TRPV1 at 4 degrees Celsius | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, Phosphatidylinositol, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LPB
 
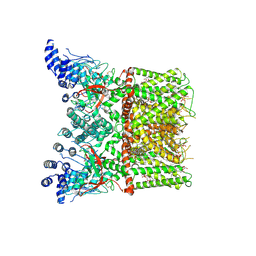 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 25 degrees Celsius | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5DEJ
 
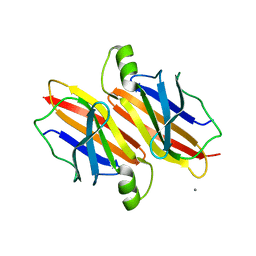 | | Transthyretin natural mutant A19D | | Descriptor: | CALCIUM ION, Transthyretin | | Authors: | Pereira, H.M, Ferreira, P, Andrade, C, Lima, C, Palhano, F, Foguel, D. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of human Transthyretin natural mutant A19D
To Be Published
|
|
7LPC
 
 | | Cryo-EM structure of full-length TRPV1 at 48 degrees Celsius | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, Phosphatidylinositol, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5AQQ
 
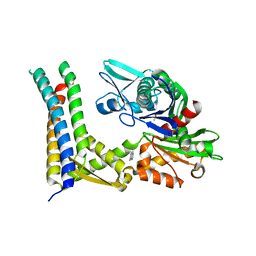 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7-methylquinazolin-4-amine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQR
 
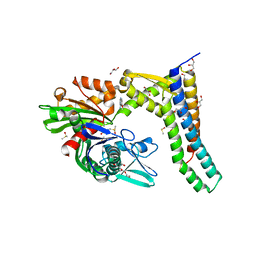 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-methoxyquinazolin-4-amine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
3BUT
 
 | | Crystal structure of protein Af_0446 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein Af_0446 | | Authors: | Bonanno, J.B, Patskovsky, Y, Ozyurt, S, Ashok, S, Zhang, F, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-03 | | Release date: | 2008-01-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of protein Af_0446 from Archaeoglobus fulgidus.
To be Published
|
|
3H4M
 
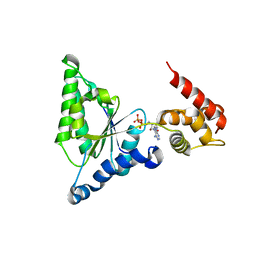 | | AAA ATPase domain of the proteasome- activating nucleotidase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Proteasome-activating nucleotidase | | Authors: | Jeffrey, P, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3E64
 
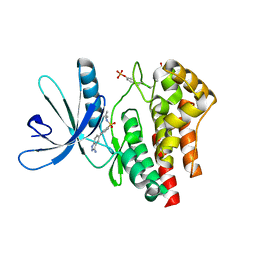 | | Fragment based discovery of JAK-2 inhibitors | | Descriptor: | 4-(3-amino-1H-indazol-5-yl)-N-tert-butylbenzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E63
 
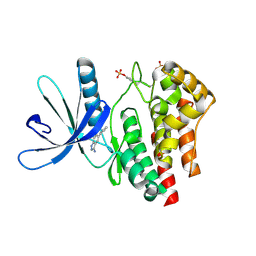 | | Fragment based discovery of JAK-2 inhibitors | | Descriptor: | 5-phenyl-1H-indazol-3-amine, Tyrosine-protein kinase JAK2 | | Authors: | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
