5I95
 
 | | Crystal Structure of Human Mitochondrial Isocitrate Dehydrogenase R140Q Mutant Homodimer bound to NADPH and alpha-Ketoglutaric acid | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Zhang, B, Jin, L, Wu, W, Jiang, F, DeLaBarre, B, Travins, J.A, Padyana, A.K. | | Deposit date: | 2016-02-19 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | AG-221, a First-in-Class Therapy Targeting Acute Myeloid Leukemia Harboring Oncogenic IDH2 Mutations.
Cancer Discov, 7, 2017
|
|
5XHE
 
 | | Crystal structure analysis of the second bromodomain of BRD2 covalently linked to b-mercaptoethanol | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, TRIETHYLENE GLYCOL | | Authors: | Padmanabhan, B, Mathur, S, Tripathi, S.K, Deshmukh, P. | | Deposit date: | 2017-04-20 | | Release date: | 2017-09-06 | | Last modified: | 2018-08-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into the crystal structure of BRD2-BD2 - phenanthridinone complex and theoretical studies on phenanthridinone analogs.
J. Biomol. Struct. Dyn., 36, 2018
|
|
6IV9
 
 | | the Cas13d binary complex | | Descriptor: | Cas13d, MAGNESIUM ION, crRNA (50-MER) | | Authors: | Zhang, B, Ye, Y.M, Ye, W.W, OuYang, S.Y. | | Deposit date: | 2018-12-02 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d.
Nat Commun, 10, 2019
|
|
5XHK
 
 | | Crystal structure of the BRD2-BD2 in complex with phenanthridinone | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, METHOXYETHANE, ... | | Authors: | Padmanabhan, B, Mathur, S, Tripathi, S, Deshmukh, P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Insights into the crystal structure of BRD2-BD2 - phenanthridinone complex and theoretical studies on phenanthridinone analogs.
J. Biomol. Struct. Dyn., 36, 2018
|
|
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
6IV8
 
 | | the selenomethionine(SeMet)-derived Cas13d binary complex | | Descriptor: | MAGNESIUM ION, RNA (51-MER), RNA (53-MER), ... | | Authors: | Zhang, B, Ye, Y.M, Ye, W.W, OuYang, S.Y. | | Deposit date: | 2018-12-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d.
Nat Commun, 10, 2019
|
|
4YZO
 
 | | Crystal Structure Analysis of Thiolase-like protein, ST0096 from Sulfolobus Tokodaii | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Padmanabhan, B, Manjula, R, Yokoyama, S, Bessho, Y. | | Deposit date: | 2015-03-25 | | Release date: | 2016-03-30 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Analysis of Thiolase-like protein, ST0096 from Sulfolobus Tokodaii
To Be Published
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5XNW
 
 | |
7XX3
 
 | | Crystal structure of human Superoxide Dismutase (SOD1) in complex with a fungal metabolite molecule, Phialomustin B (PB) | | Descriptor: | (2~{E},4~{E},6~{S})-4,6-dimethyldeca-2,4-dienoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Padmanabhan, B, Unni, S. | | Deposit date: | 2022-05-28 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phialomustin-B a fungal metabolite isolated from Phialophora mustea modulates Superoxide Dismutase 1 (SOD1) aggregation: Therapeutic potential in Amyotrophic lateral sclerosis (ALS)
To Be Published
|
|
7YZ9
 
 | | Structure of catalytic domain of Rv1625c bound to nanobody NB4 | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, Adenylate cyclase, GLYCEROL, ... | | Authors: | Khanppnavar, B, Mehta, V.J, Iype, T, Korkhov, V.M. | | Deposit date: | 2022-02-19 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of Mycobacterium tuberculosis Cya, an evolutionary ancestor of the mammalian membrane adenylyl cyclases.
Elife, 11, 2022
|
|
8H1I
 
 | | Crystal structure of PlyGRCS, a bacteriophage Endolysin in complex with Cold shock protein C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cold shock-like protein CspC, ... | | Authors: | Padmanabhan, B, Gopinatha, K, Mandal, M, Saranya, G, Sudhagar, B. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PlyGRCS, a bacteriophage Endolysin in complex with Cold shock protein C
To Be Published
|
|
7ZH0
 
 | |
7ZH6
 
 | |
5YTO
 
 | |
7ZHA
 
 | |
8I16
 
 | | Crystal structure of the selenomethionine (SeMet)-derived Cas12g (D513A) mutant | | Descriptor: | Cas12g, ZINC ION | | Authors: | Zhang, B, Chen, J, Ye, Y.M, OuYang, S.Y. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural transitions upon guide RNA binding and their importance in Cas12g-mediated RNA cleavage.
Plos Genet., 19, 2023
|
|
5ZIK
 
 | | Crystal structure of Ketopantoate reductase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Probable 2-dehydropantoate 2-reductase, SULFATE ION | | Authors: | Khanppnavar, B, Datta, S. | | Deposit date: | 2018-03-16 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Genome-wide survey and crystallographic analysis suggests a role for both horizontal gene transfer and duplication in pantothenate biosynthesis pathways.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
8FET
 
 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEV
 
 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) and dihydroquercetin complex | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FIO
 
 | | Hypothetical anthocyanidin reductase from Sorghum bicolor-NADP(H) and naringenin complex | | Descriptor: | Epimerase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NARINGENIN | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEW
 
 | | Flavanone 4-Reductase from Sorghum bicolor-naringenin complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NARINGENIN, SULFATE ION | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FIP
 
 | | Hypothetical anthocyanidin reducatase from Sorghum bicolor- NADP+ complex | | Descriptor: | Epimerase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEU
 
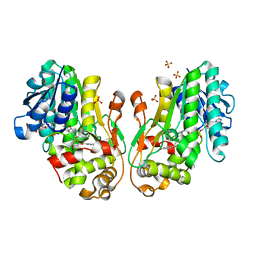 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) and naringenin complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NARINGENIN, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
5TLQ
 
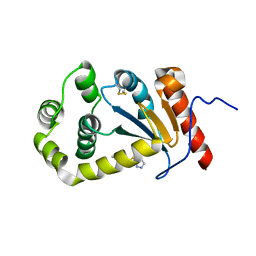 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
