6WNF
 
 | |
8S1X
 
 | |
7BBL
 
 | |
8HSI
 
 | | Cryo-EM structure of human TMEM87A, PE-bound | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8HTT
 
 | | Cryo-EM structure of human TMEM87A, gluconate-bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-gluconic acid, Transmembrane protein 87A,EGFP, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8KB4
 
 | | Cryo-EM structure of human TMEM87A A308M | | Descriptor: | (9R,12R)-15-amino-12-hydroxy-6,12-dioxo-7,11,13-trioxa-12lambda~5~-phosphapentadecan-9-yl undecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 87A,EGFP | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
6DCM
 
 | |
5EY0
 
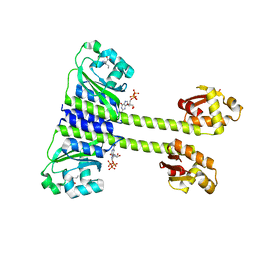 | | Crystal structure of CodY from Staphylococcus aureus with GTP and Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, GUANOSINE-5'-TRIPHOSPHATE, ISOLEUCINE | | Authors: | Han, A, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
5EY2
 
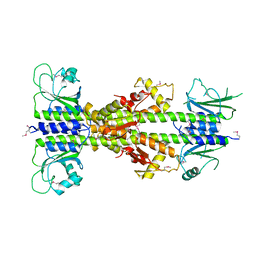 | | Crystal structure of CodY from Bacillus cereus | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY | | Authors: | Han, A, Lee, W.C, Son, J, Kim, S.H, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
5EY1
 
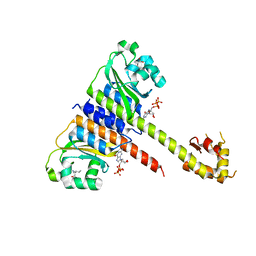 | | Crystal structure of CodY from Staphylococcus aureus with GTP and Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, GUANOSINE-5'-TRIPHOSPHATE, ISOLEUCINE | | Authors: | Han, A, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
5FA9
 
 | | Bifunctional Methionine Sulfoxide Reductase AB (MsrAB) from Treponema denticola | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptide methionine sulfoxide reductase MsrA | | Authors: | Han, A, Son, J, Kim, H.-Y, Hwang, K.Y. | | Deposit date: | 2015-12-11 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Essential Role of the Linker Region in the Higher Catalytic Efficiency of a Bifunctional MsrA-MsrB Fusion Protein
Biochemistry, 55, 2016
|
|
1N6J
 
 | | Structural basis of sequence-specific recruitment of histone deacetylases by Myocyte Enhancer Factor-2 | | Descriptor: | 5'-D(*AP*GP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*CP*T)-3', Calcineurin-binding protein Cabin 1, ... | | Authors: | Han, A, Pan, F, Stroud, J.C, Youn, H.D, Liu, J.O, Chen, L. | | Deposit date: | 2002-11-11 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2
Nature, 422, 2003
|
|
3LST
 
 | | Crystal Structure of CalO1, Methyltransferase in Calicheamicin Biosynthesis, SAH bound form | | Descriptor: | 1,2-ETHANEDIOL, CalO1 Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of CalO1: a putative orsellinic acid methyltransferase in the calicheamicin-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OTI
 
 | | Crystal Structure of CalG3, Calicheamicin Glycostyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CHLORIDE ION, CalG3, Calicheamicin T0, ... | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTH
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP and calicheamicin alpha3I bound form | | Descriptor: | CalG1, Calicheamicin alpha3I, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTG
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form | | Descriptor: | CHLORIDE ION, CalG1, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PKP
 
 | | Q83S Variant of S. Enterica RmlA with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Glucose-1-phosphate thymidylyltransferase, MAGNESIUM ION | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
3PKQ
 
 | | Q83D Variant of S. Enterica RmlA with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
7L73
 
 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to ERK5-IN-1 | | Descriptor: | 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-12-25 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LEJ
 
 | | Crystal structure of the second bromodomain (BD2) of human BRDT bound to Volasertib | | Descriptor: | Bromodomain testis-specific protein, N-{trans-4-[4-(cyclopropylmethyl)piperazin-1-yl]cyclohexyl}-4-{[(7R)-7-ethyl-5-methyl-8-(1-methylethyl)-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxybenzamide | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LEK
 
 | | Crystal structure of the second bromodomain (BD2) of human BRDT bound to ERK5-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein, ... | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LEL
 
 | |
7LEM
 
 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to LRRK2-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LRK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-093 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{S})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LP0
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-077 | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorol-2-methyl-5-[[(3~{R})-1-methylpiperidin-3-yl]amino]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
