8F17
 
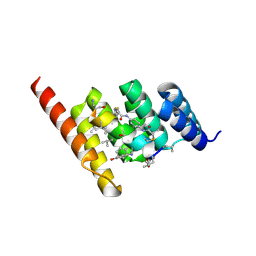 | | Structure of the STUB1 TPR domain in complex with H204, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
3HU7
 
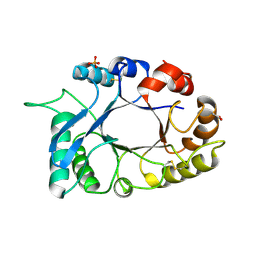 | | Structural characterization and binding studies of a plant pathogenesis related protein heamanthin from haemanthus multiflorus reveal its dual inhibitory effects against xylanase and alpha-amylase | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-06-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
8F15
 
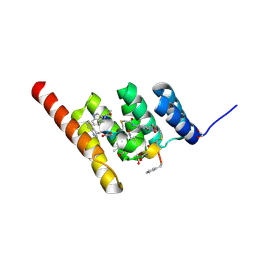 | | Structure of the STUB1 TPR domain in complex with H202, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8F0Z
 
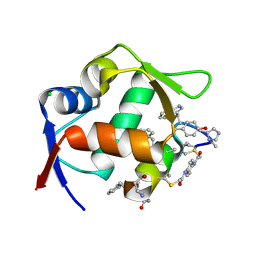 | | Structure of the MDM2 P53 binding domain in complex with H101, an all-D Helicon Polypeptide | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, H101, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8F16
 
 | | Structure of the STUB1 TPR domain in complex with H203, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8F14
 
 | | Structure of the STUB1 TPR domain in complex with H201, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8F10
 
 | | Structure of the MDM2 P53 binding domain in complex with H102, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
1OWQ
 
 | | Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, signal processing protein | | Authors: | Kumar, J, Sharma, S, Jasti, J, Bhushan, A, Singh, T.P. | | Deposit date: | 2003-03-29 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution
To be Published
|
|
6JCE
 
 | | NMR solution and X-ray crystal structures of a DNA containing both right-and left-handed parallel-stranded G-quadruplexes | | Descriptor: | 29-mer DNA | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
6S5I
 
 | |
2MAY
 
 | |
2M91
 
 | |
2M90
 
 | |
2M8Y
 
 | | Structure of d[CGCGAAGCATTCGCG] hairpin | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*GP*CP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Lim, K.W, Phan, A.T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA quadruplex-duplex junction formation.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3CG9
 
 | | Crystal structure of the complex of peptidoglycan recognition protein with methyloxane-2,3,4,5-tetrol at 2.9 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein, alpha-L-rhamnopyranose | | Authors: | Sharma, P, Kaur, A, Singh, N, Sharma, S, Bhushan, A, Pathak, K.M.L, Kaur, P, Singh, T.P. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with methyoxane-2,3,4,5-tetrol at 2.9 A resolution
To be Published
|
|
2MB3
 
 | | Solution structure of an intramolecular (3+1) human telomeric G-quadruplex bound to a telomestatin derivative | | Descriptor: | (12S,27S)-12,27-bis(4-aminobutyl)-4,30-dimethyl-3,7,14,18,22,29-hexaoxa-11,26,31,32,33,34,35,36-octaazaheptacyclo[26.2. 1.1~2,5~.1~6,9~.1~13,16~.1~17,20~.1~21,24~]hexatriaconta-1(30),2(36),4,6(35),8,13(34),15,17(33),19,21(32),23,28(31)-dode caene-10,25-dione, DNA_(5'-D(*TP*TP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*A)-3') | | Authors: | Chung, W.J, Heddi, B, Tera, M, Iida, K, Nagasawa, K, Phan, A.T. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular (3 + 1) human telomeric g-quadruplex bound to a telomestatin derivative.
J.Am.Chem.Soc., 135, 2013
|
|
2MB4
 
 | | Solution structure of a stacked dimeric G-quadruplex formed by a segment of the human CEB1 minisatellite | | Descriptor: | DNA_(5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*TP*GP*G)-3') | | Authors: | Adrian, M, Ang, D.J, Lech, C, Heddi, B, Nicolas, A, Phan, A.T. | | Deposit date: | 2013-07-25 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Conformational Dynamics of a Stacked Dimeric G-Quadruplex Formed by the Human CEB1 Minisatellite.
J.Am.Chem.Soc., 136, 2014
|
|
2M93
 
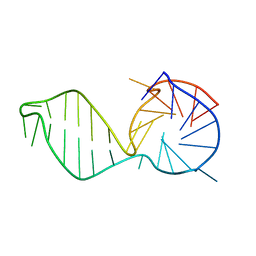 | |
2MGN
 
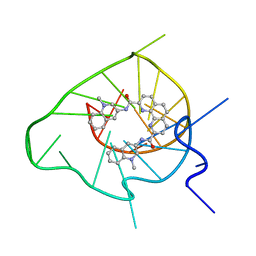 | | Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC3 | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3', N2,N9-bis(1-methylquinolin-3-yl)-1,10-phenanthroline-2,9-dicarboxamide | | Authors: | Chung, W.J, Heddi, B, Hamon, F, Teulade-Fichou, M.P, Phan, A.T. | | Deposit date: | 2013-11-01 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a G-quadruplex Bound to the Bisquinolinium Compound Phen-DC3.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6SPF
 
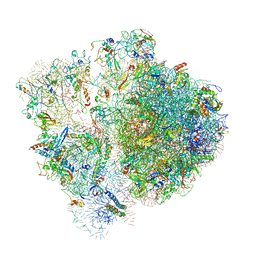 | | Pseudomonas aeruginosa 70s ribosome from an aminoglycoside resistant clinical isolate | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7CV3
 
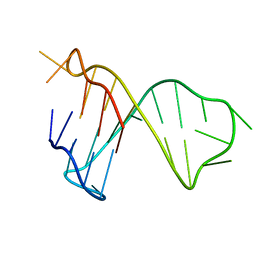 | | Quadruplex-duplex hybrid structure in the PIM1 gene, Form 1 | | Descriptor: | PIM1 promoter, Form 1 | | Authors: | Winnerdy, F.R, Tan, D.J.Y, Lim, K.W, Phan, A.T. | | Deposit date: | 2020-08-25 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Coexistence of two quadruplex-duplex hybrids in the PIM1 gene.
Nucleic Acids Res., 48, 2020
|
|
2MWZ
 
 | |
6SPE
 
 | | Pseudomonas aeruginosa 30s ribosome from a clinical isolate | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1CY1
 
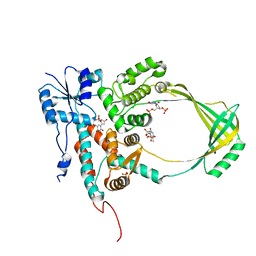 | | COMPLEX OF E.COLI DNA TOPOISOMERASE I WITH 5'PTPTPT | | Descriptor: | DNA TOPOISOMERASE I, PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, ... | | Authors: | Feinberg, H, Changela, A, Mondragon, A. | | Deposit date: | 1999-08-31 | | Release date: | 2000-03-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-nucleotide interactions in E. coli DNA topoisomerase I.
Nat.Struct.Biol., 6, 1999
|
|
6SPC
 
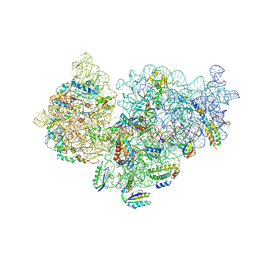 | | Pseudomonas aeruginosa 30s ribosome from an aminoglycoside resistant clinical isolate | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
