2WLU
 
 | | Iron-bound crystal structure of Streptococcus pyogenes Dpr | | Descriptor: | DPS-LIKE PEROXIDE RESISTANCE PROTEIN, FE (III) ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of Streptococcus Pyogenes Dpr Reveal a Dodecameric Iron-Binding Protein with a Ferroxidase Site.
J.Biol.Inorg.Chem., 15, 2010
|
|
5FSZ
 
 | |
5FPG
 
 | | Crystal structure of human tankyrase 2 in complex with TA-92 | | Descriptor: | 4-[3-(8-METHYL-4-OXO-3,4-DIHYDROQUINAZOLIN-2- YL)PROPANAMIDO]-N-(QUINOLIN-8-YL)BENZAMIDE, TANKYRASE-2, ZINC ION | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Synthesis and Evaluation of Novel Dual- Binding Inhibitors of the Tankyrases and Wnt Signalling.
To be Published
|
|
5FSX
 
 | |
5FSU
 
 | |
5FSY
 
 | | Crystal structure of Trypanosoma brucei macrodomain in complex with ADP-ribose | | Descriptor: | MACRODOMAIN, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2016-01-08 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Proximal Adp-Ribose Hydrolysis in Trypanosomatids is Catalyzed by a Macrodomain.
Sci.Rep., 6, 2016
|
|
5FPF
 
 | | Crystal structure of human tankyrase 2 in complex with TA-91 | | Descriptor: | 4-[3-(4-oxo-3,4-dihydroquinazolin-2- yl)propanamido]-N-(quinolin-8-yl)benzamide, TANKYRASE-2, ZINC ION | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, Synthesis and Evaluation of Novel Dual- Binding Inhibitors of the Tankyrases and Wnt Signalling.
To be Published
|
|
5FSV
 
 | |
5ADQ
 
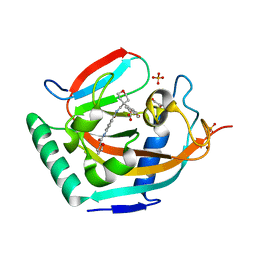 | | Crystal structure of human tankyrase 2 in complex with JW55 | | Descriptor: | BICARBONATE ION, GLYCEROL, N-(4-(((4-(4-methoxyphenyl)oxan-4- yl)methyl)carbamoyl)phenyl)furan-2-carboxamide, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development and Structural Analysis of Adenosine Site Binding Tankyrase Inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5ADR
 
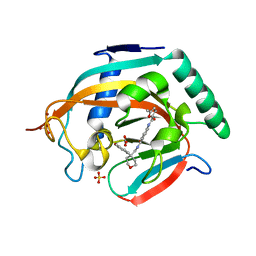 | | Crystal structure of human tankyrase 2 in complex with OD38 | | Descriptor: | BICARBONATE ION, N-(4-(((4-(4-methoxyphenyl)oxan-4- yl)methyl)carbamoyl)phenyl)-5-methylfuran-2-carboxamide, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development and Structural Analysis of Adenosine Site Binding Tankyrase Inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5ADT
 
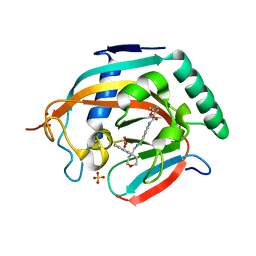 | | Crystal structure of human tankyrase 2 in complex with OD73 | | Descriptor: | BICARBONATE ION, N-[3-chloranyl-4-[[4-(4-methoxyphenyl)oxan-4-yl]methylcarbamoyl]phenyl]-2-methyl-1,3-oxazole-5-carboxamide, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development and Structural Analysis of Adenosine Site Binding Tankyrase Inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8BX6
 
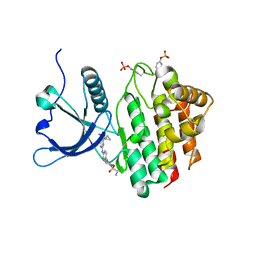 | |
8BXH
 
 | |
8BXC
 
 | |
8BX9
 
 | |
8BA4
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BA3
 
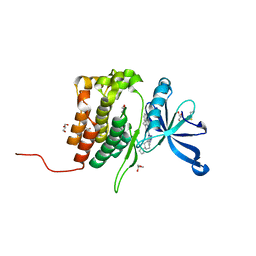 | | Crystal structure of JAK2 JH2 in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B99
 
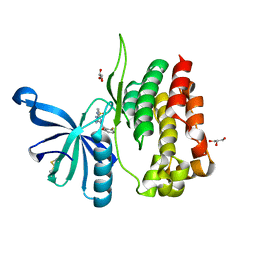 | | Crystal structure of JAK2 JH2-V617F in complex with JNJ-7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8U
 
 | |
8B9H
 
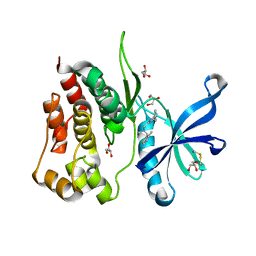 | | Crystal structure of JAK2 JH2 in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B9E
 
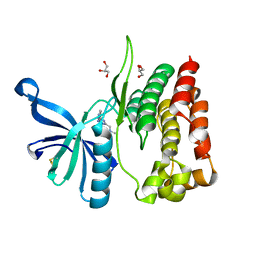 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BAK
 
 | |
8BAB
 
 | | Crystal structure of JAK2 JH2-V617F in complex with CB76 | | Descriptor: | 6-[(1-methylimidazol-2-yl)sulfanylmethyl]-~{N}4-(3-methylphenyl)-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8N
 
 | |
8BA2
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A1 | | Descriptor: | 6-[[(5-bromanylthiophen-2-yl)methyl-methyl-amino]methyl]-~{N}4-(4-methylphenyl)-1,3,5-triazine-2,4-diamine, ACETATE ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
