2KHM
 
 | | Structure of the C-terminal non-repetitive domain of the spider dragline silk protein ADF-3 | | Descriptor: | Fibroin-3 | | Authors: | Hagn, F.X, Eisoldt, L, Hardy, J.G, Vendrely, C, Coles, M, Scheibel, T, Kessler, H. | | Deposit date: | 2009-04-09 | | Release date: | 2010-04-14 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | A conserved spider silk domain acts as a molecular switch that controls fibre assembly
Nature, 465, 2010
|
|
7BGH
 
 | |
6F46
 
 | |
2M07
 
 | | NMR structure of OmpX in DPC micelles | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M06
 
 | | NMR structure of OmpX in phopspholipid nanodiscs | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
6Z0G
 
 | | Structure of TREM2 transmembrane helix in DPC micelles | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Steiner, A, Schlepkow, K, Brunner, B, Steiner, H, Haass, C, Hagn, F. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | gamma-Secretase cleavage of the Alzheimer risk factor TREM2 is determined by its intrinsic structural dynamics.
Embo J., 39, 2020
|
|
6Z0I
 
 | | Structure of the TREM2 transmembrane helix in complex with DAP12 in DPC micelles | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Steiner, A, Schlepkow, K, Brunner, B, Steiner, H, Haass, C, Hagn, F. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | gamma-Secretase cleavage of the Alzheimer risk factor TREM2 is determined by its intrinsic structural dynamics.
Embo J., 2020
|
|
6Z0H
 
 | | Structure of TREM2 transmembrane helix K186A variant in DPC micelles | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Steiner, A, Schlepkow, K, Brunner, B, Steiner, H, Haass, C, Hagn, F. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | gamma-Secretase cleavage of the Alzheimer risk factor TREM2 is determined by its intrinsic structural dynamics.
Embo J., 39, 2020
|
|
2AKP
 
 | | Hsp90 Delta24-N210 mutant | | Descriptor: | ATP-dependent molecular chaperone HSP82 | | Authors: | Richter, K, Moser, S, Hagn, F, Friedrich, R, Hainzl, O, Heller, M, Schlee, S, Kessler, H, Reinstein, J, Buchner, J. | | Deposit date: | 2005-08-03 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Intrinsic inhibition of the Hsp90 ATPase activity.
J.Biol.Chem., 281, 2006
|
|
7OFM
 
 | |
7OFO
 
 | |
5JS8
 
 | | Structural Model of a Protein alpha subunit in complex with GDP obtained with SAXS and NMR residual couplings | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | Authors: | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | Deposit date: | 2016-05-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JS7
 
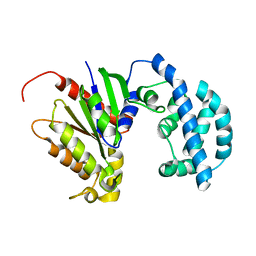 | | Structural model of a apo G-protein alpha subunit determined with NMR residual dipolar couplings and SAXS | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | Authors: | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | Deposit date: | 2016-05-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3UQ3
 
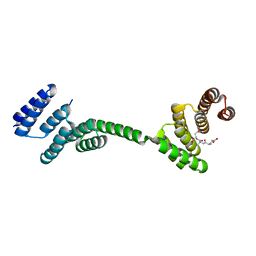 | | TPR2AB-domain:pHSP90-complex of yeast Sti1 | | Descriptor: | Heat shock protein, Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
3UPV
 
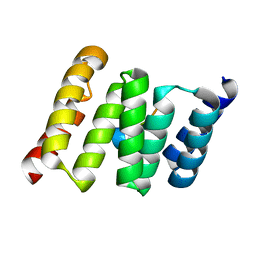 | | TPR2B-domain:pHsp70-complex of yeast Sti1 | | Descriptor: | Heat shock protein SSA4, Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
7L0R
 
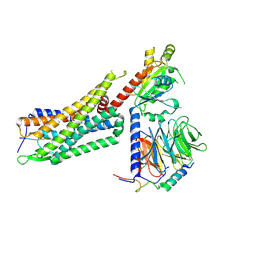 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0P
 
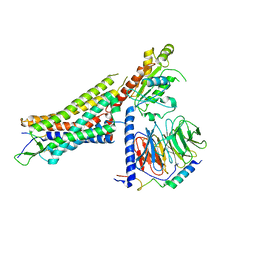 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0S
 
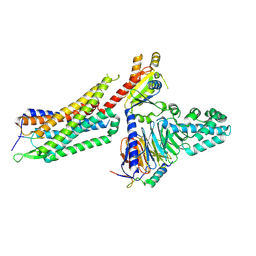 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2LLW
 
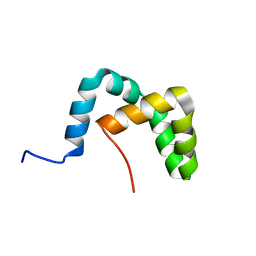 | | Solution structure of the yeast Sti1 DP2 domain | | Descriptor: | Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
2LLV
 
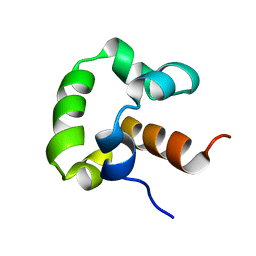 | | Solution structure of the yeast Sti1 DP1 domain | | Descriptor: | Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
