1OPY
 
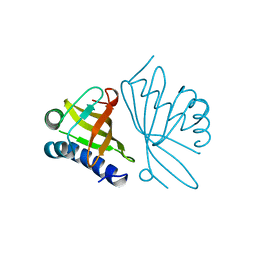 | | KSI | | Descriptor: | DELTA5-3-KETOSTEROID IOSMERASE | | Authors: | Kim, S.-W, Cha, S.-S, Cho, H.-S, Kim, J.-S, Ha, N.-C, Cho, M.-J, Choi, K.-Y, Oh, B.-H. | | Deposit date: | 1997-05-23 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures of delta5-3-ketosteroid isomerase with and without a reaction intermediate analogue.
Biochemistry, 36, 1997
|
|
1CVM
 
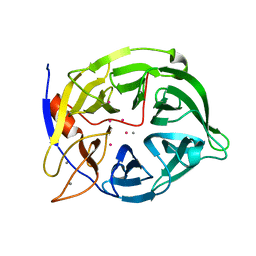 | | CADMIUM INHIBITED CRYSTAL STRUCTURE OF PHYTASE FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | CADMIUM ION, CALCIUM ION, PHYTASE | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-24 | | Release date: | 2000-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
6KOD
 
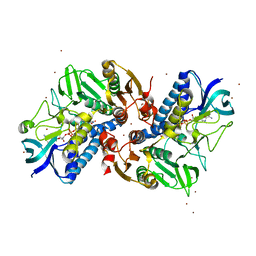 | |
6KYY
 
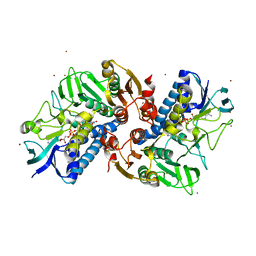 | |
6IY0
 
 | |
6K25
 
 | |
6K22
 
 | |
7FB9
 
 | | Crystal Structure of Human Cu, Zn Superoxide Dismutase (SOD1) | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Baek, Y, Ha, N.-C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of the overoxidized Cu/Zn-superoxide dismutase in ROS-induced ALS filament formation.
Commun Biol, 5, 2022
|
|
6KGY
 
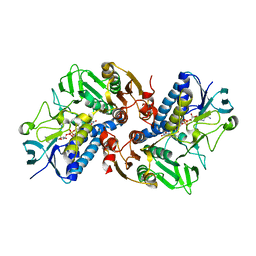 | | HOCl-induced flavoprotein disulfide reductase RclA from Escherichia coli | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pyridine nucleotide-disulphide oxidoreductase dimerisation region | | Authors: | Baek, Y, Ha, N.-C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of the hypochlorous acid-induced flavoprotein RclA fromEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
7FB6
 
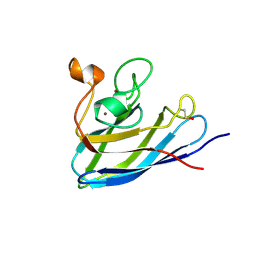 | | C57D/C146D mutant of Human Cu, Zn Superoxide Dismutase (SOD1) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Baek, Y, Ha, N.-C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the overoxidized Cu/Zn-superoxide dismutase in ROS-induced ALS filament formation.
Commun Biol, 5, 2022
|
|
1OCL
 
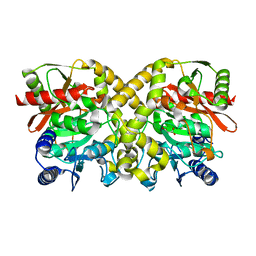 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COMPLEXED WITH MALONATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, MALONIC ACID | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1OCK
 
 | |
1OCM
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COVALENTLY COMPLEXED WITH PYROPHOSPHATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, PYROPHOSPHATE 2- | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
7DNP
 
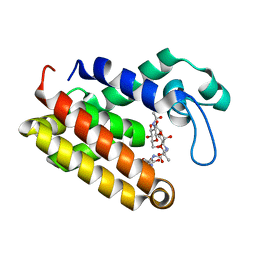 | | Structure of Brucella abortus SagA | | Descriptor: | (2R)-2-[[(2S)-2-[[(2R)-2-[(2R,3S,4R,5R,6S)-5-acetamido-3-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-(hydroxymethyl)-6-oxidanyl-oxan-4-yl]oxypropanoyl]amino]propanoyl]amino]pentanedioic acid, Secretion activator protein, hypothetical | | Authors: | Hyun, Y, Ha, N.-C. | | Deposit date: | 2020-12-10 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of the Autolysin SagA in the Type IV Secretion System of Brucella abortus .
Mol.Cells, 44, 2021
|
|
7DPY
 
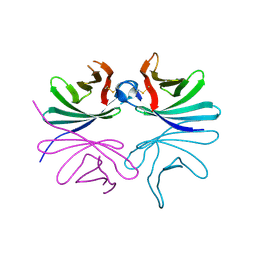 | | Structure of Brucella abortus PhiA | | Descriptor: | Brucella Abortus PhiA | | Authors: | Hyun, Y, Ha, N.-C. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the Autolysin SagA in the Type IV Secretion System of Brucella abortus .
Mol.Cells, 44, 2021
|
|
7DTG
 
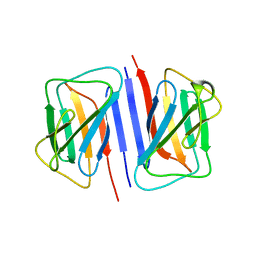 | |
7DXN
 
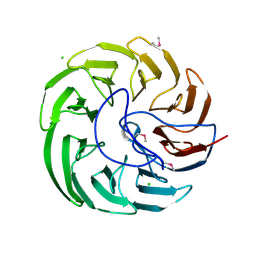 | | Plant growth-promoting factor YxaL from Bacillus velezensis | | Descriptor: | CHLORIDE ION, Membrane associated protein kinase with beta-propeller domain, pyrrolo-quinoline quinone beta-propeller repeat | | Authors: | Baek, Y, Ha, N.-C. | | Deposit date: | 2021-01-19 | | Release date: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the plant growth-promoting factor YxaL from the rhizobacterium Bacillus velezensis and its application to protein engineering.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7EQ5
 
 | | Plant growth-promoting factor YxaL mutant from Bacillus velezensis - T175W/W215G | | Descriptor: | Membrane associated protein kinase with beta-propeller domain, pyrrolo-quinoline quinone beta-propeller repeat | | Authors: | Kim, J, Ha, N.-C. | | Deposit date: | 2021-04-30 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the plant growth-promoting factor YxaL from the rhizobacterium Bacillus velezensis and its application to protein engineering.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7DOG
 
 | |
7ERQ
 
 | |
7ERP
 
 | |
7FDF
 
 | |
7EVF
 
 | |
1GS3
 
 | |
