5AZB
 
 | | Crystal structure of Escherichia coli Lgt in complex with phosphatidylglycerol and the inhibitor palmitic acid | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PALMITIC ACID, Prolipoprotein diacylglyceryl transferase, ... | | Authors: | Zhang, X.C, Mao, G, Zhao, Y. | | Deposit date: | 2015-09-30 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of E. coli lipoprotein diacylglyceryl transferase
Nat Commun, 7, 2016
|
|
5AZC
 
 | |
4GJE
 
 | |
7F8W
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7FFO
 
 | | Cryo-EM structure of VEEV VLP at the 5-fold axes | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFQ
 
 | | Cryo-EM structure of VEEV VLP at the 2-fold axes | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7F8V
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7FFE
 
 | | Cryo-EM structure of VEEV VLP | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFL
 
 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 2-fold axes | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFN
 
 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 5-fold axes | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
4GJF
 
 | |
4GJG
 
 | |
5XBM
 
 | | Structure of SCARB2-JL2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
7XGD
 
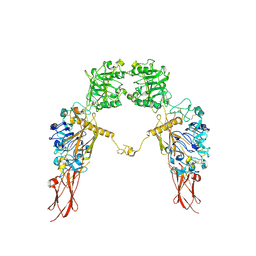 | | Cryo-EM structure of Apo-IGF1R map 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor 1 receptor | | Authors: | Zhang, X, Wu, C. | | Deposit date: | 2022-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of Apo-IGF1R
To Be Published
|
|
2GS7
 
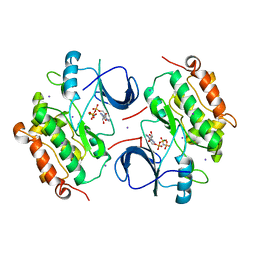 | | Crystal Structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
3J9D
 
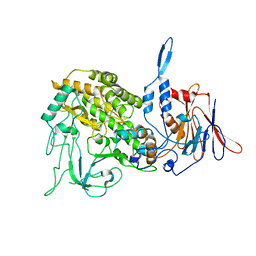 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | Outer capsid protein VP2, ZINC ION | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3J9E
 
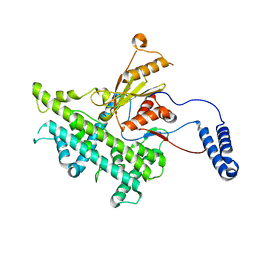 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | VP5 | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1E32
 
 | | Structure of the N-Terminal domain and the D1 AAA domain of membrane fusion ATPase p97 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, P97 | | Authors: | Zhang, X, Shaw, A, Bates, P.A, Gorman, M.A, Kondo, H, Dokurno, P, Leonard M, G, Sternberg, J.E, Freemont, P.S. | | Deposit date: | 2000-06-05 | | Release date: | 2001-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Aaa ATPase P97
Mol.Cell, 6, 2000
|
|
1J04
 
 | | Structural mechanism of enzyme mistargeting in hereditary kidney stone disease in vitro | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, alanine--glyoxylate aminotransferase | | Authors: | Zhang, X, Djordjevic, S, Bartlam, M, Ye, S, Rao, Z, Danpure, C.J. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural implications of a G170R mutation of alanine:glyoxylate aminotransferase that is associated with peroxisome-to-mitochondrion mistargeting.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1UON
 
 | | REOVIRUS POLYMERASE LAMBDA-3 LOCALIZED BY ELECTRON CRYOMICROSCOPY OF VIRIONS AT 7.6-A RESOLUTION | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 5'-R(*GP*GP*GP*GP*GP*)-3', 5'-R(*UP*AP*GP*CP*CP*CP*CP*CP*)-3', ... | | Authors: | Zhang, X, Walker, S.B, Chipman, P.R, Nibert, M.L, Baker, T.S. | | Deposit date: | 2003-09-21 | | Release date: | 2003-11-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Reovirus Polymerase Lambda3 Localized by Cryo-Electron Microscopy of Virions at a Resolution of 7.6 A
Nat.Struct.Biol., 10, 2003
|
|
1EUB
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED TO A POTENT NON-PEPTIDIC SULFONAMIDE INHIBITOR | | Descriptor: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | Authors: | Zhang, X, Gonnella, N.C, Koehn, J, Pathak, N, Ganu, V, Melton, R, Parker, D, Hu, S.I, Nam, K.Y. | | Deposit date: | 2000-04-14 | | Release date: | 2001-04-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human collagenase-3 (MMP-13) complexed to a potent non-peptidic sulfonamide inhibitor: binding comparison with stromelysin-1 and collagenase-1.
J.Mol.Biol., 301, 2000
|
|
6IMT
 
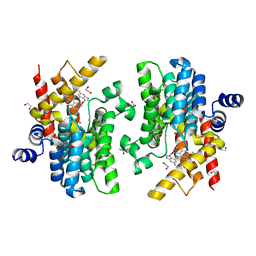 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(6-fluoro-1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
5YT1
 
 | |
8JT4
 
 | |
7YOB
 
 | |
