4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6IG0
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1, ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTR1, MAGNESIUM ION, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
4JPP
 
 | | Bacteriophage phiX174 H protein residues 143-282 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
4PEK
 
 | | Crystal structure of a computationally designed retro-aldolase, RA114.3 | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4PEJ
 
 | | Crystal structure of a computationally designed retro-aldolase, RA110.4 (Cys free) | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
3UUO
 
 | | The discovery of potent, selectivity, and orally bioavailable pyrozoloquinolines as PDE10 inhibitors for the treatment of Schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(piperazin-1-yl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ho, G.D, Yang, S, Smotryski, J, Bercovici, A, Nechuta, T, Smith, E.M, McElroy, W, Tan, Z, Tulshian, D, Mckittrick, B, Greenlee, W.J, Hruza, A, Xiao, L, Rindgen, D, Guzzi, M, Zhang, X, Bleickardt, C, Mullins, D, Hodgson, R. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The discovery of potent, selective, and orally active pyrazoloquinolines as PDE10A inhibitors for the treatment of Schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4LDT
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBCH5B~Ub | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin, ... | | Authors: | Wiener, R, DiBello, A.T, Lombardi, P.M, Guzzo, C.M, Zhang, X, Matunis, M.J, Wolberger, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2AIY
 
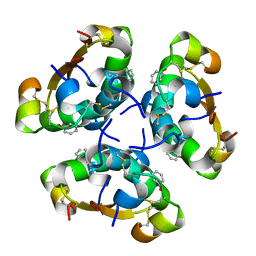 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 20 STRUCTURES | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-28 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
4PX7
 
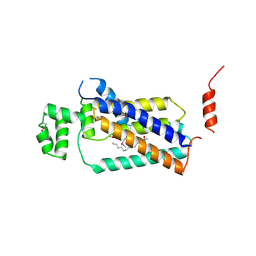 | | Crystal structure of lipid phosphatase E. coli PgpB | | Descriptor: | GLYCEROL, LAURYL DIMETHYLAMINE-N-OXIDE, Phosphatidylglycerophosphatase | | Authors: | Fan, J, Jiang, D, Zhao, Y, Zhang, X.C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-05-28 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2F0X
 
 | | Crystal structure and function of human thioesterase superfamily member 2(THEM2) | | Descriptor: | SULFATE ION, Thioesterase superfamily member 2 | | Authors: | Cheng, Z, Song, F, Shan, X, Wang, Y, Wei, Z, Gong, W. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human thioesterase superfamily member 2
Biochem.Biophys.Res.Commun., 349, 2006
|
|
6KA4
 
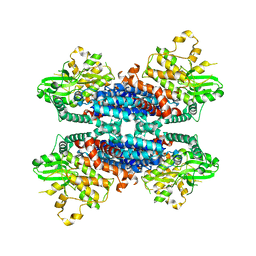 | | Cryo-EM structure of the AtMLKL3 tetramer | | Descriptor: | F22L4.1 protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-06-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
4M8M
 
 | |
4LT5
 
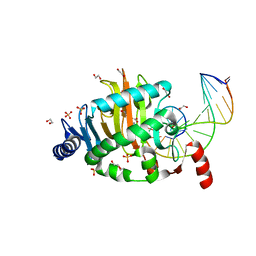 | | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA, MANGANESE (II) ION, ... | | Authors: | Hashimoto, H, Pais, J.E, Zhang, X, Saleh, L, Fu, Z.Q, Dai, N, Correa, I.R, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.
Nature, 506, 2014
|
|
4M9E
 
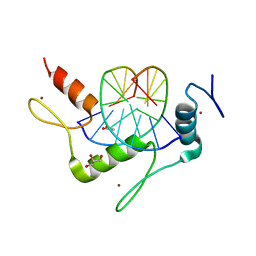 | | Structure of Klf4 zinc finger DNA binding domain in complex with methylated DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*AP*GP*GP*(5CM)P*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*(5CM)P*GP*CP*CP*TP*C)-3'), ... | | Authors: | Liu, Y, Olanrewaju, Y.O, Blumenthal, R.M, Zhang, X, Cheng, X. | | Deposit date: | 2013-08-14 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for Klf4 recognition of methylated DNA.
Nucleic Acids Res., 42, 2014
|
|
1F3Y
 
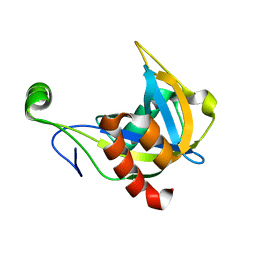 | | SOLUTION STRUCTURE OF THE NUDIX ENZYME DIADENOSINE TETRAPHOSPHATE HYDROLASE FROM LUPINUS ANGUSTIFOLIUS L. | | Descriptor: | DIADENOSINE 5',5'''-P1,P4-TETRAPHOSPHATE HYDROLASE | | Authors: | Swarbrick, J.D, Bashtannyk, T, Maksel, D, Zhang, X.R, Blackburn, G.M, Gayler, K.R, Gooley, P.R. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the Nudix enzyme diadenosine tetraphosphate hydrolase from Lupinus angustifolius L.
J.Mol.Biol., 302, 2000
|
|
4MGG
 
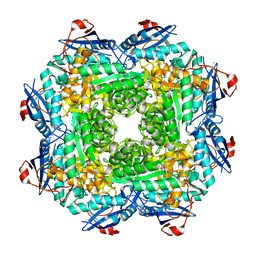 | | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Muconate lactonizing enzyme, ... | | Authors: | Vetting, M.W, Zhang, X, Wasserman, S.R, Morisco, L.L, Sojitra, S, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121
To be Published
|
|
2PJH
 
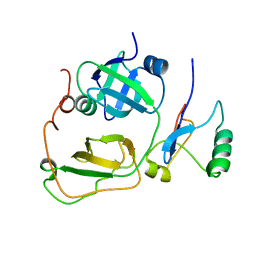 | | Strctural Model of the p97 N domain- npl4 UBD complex | | Descriptor: | Nuclear protein localization protein 4 homolog, Transitional endoplasmic reticulum ATPase | | Authors: | Isaacson, R, Pye, V.E, Simpson, S, Meyer, H.H, Zhang, X, Freemont, P. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Detailed structural insights into the p97-Npl4-Ufd1 interface.
J.Biol.Chem., 282, 2007
|
|
1LFC
 
 | | BOVINE LACTOFERRICIN (LFCINB), NMR, 20 STRUCTURES | | Descriptor: | LACTOFERRICIN | | Authors: | Hwang, P.M, Zhou, N, Shan, X, Arrowsmith, C.H, Vogel, H.J. | | Deposit date: | 1998-06-24 | | Release date: | 1998-11-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of lactoferricin B, an antimicrobial peptide derived from bovine lactoferrin.
Biochemistry, 37, 1998
|
|
2M5L
 
 | |
